Chromatin Formation and Remodeling
The most basic form of compaction of our DNA is the nucleosome as shown in the figure. DNA (blue) is wrapped around a histone octamer (grey) 1.7 times. This compact form is necessary to fit in a cell, but it makes the DNA inaccessible for proteins.
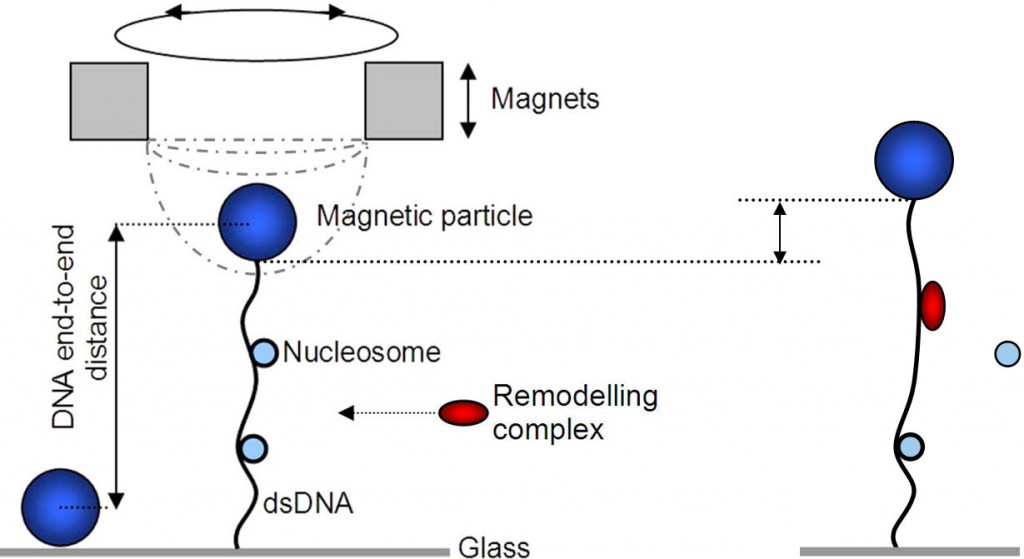
While many proteins are involved in the assembly and (re)positioning of nucleosomes, the dynamics of protein-assisted nucleosome formation are not well understood. We want to study the assembly of nucleosomes as well as the effect of remodelers on DNA with nucleosomes using single molecule techniques like magnetic tweezer and nanopore experiments. In a magnetic tweezer a piece of DNA with nucleosomes is at one end attached to a glass surface and at the other end to a magnetic bead on which we pull with a magnet. Using this technique one can monitor in real time the length with a precision of a few nanometers. Also the stretching force can be measured and changed.
We study nucleosome assembly under biologically relevant conditions mediated by the chaperone NAP1 (nucleosome assembly protein). The number of assembled nucleosomes on each of the DNA molecules can than be estimated based on the change in supercoiling density and end-to-end length.
To make important processes like transcription possible, remodeling proteins have to remove or replace the nucleosomes. Starting from the naturally formed nucleosomes we want to study the function of remodelers. One technique for this is the magnetic tweezer as explained before. Another way is to pull the entire construct of DNA, nucleosomes and remodelers through a nanopore up to the point where a remodeler sits to see its effect.
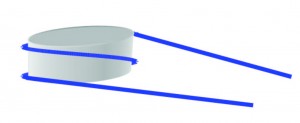
Nucleosome: DNA wrapped around histone octamer
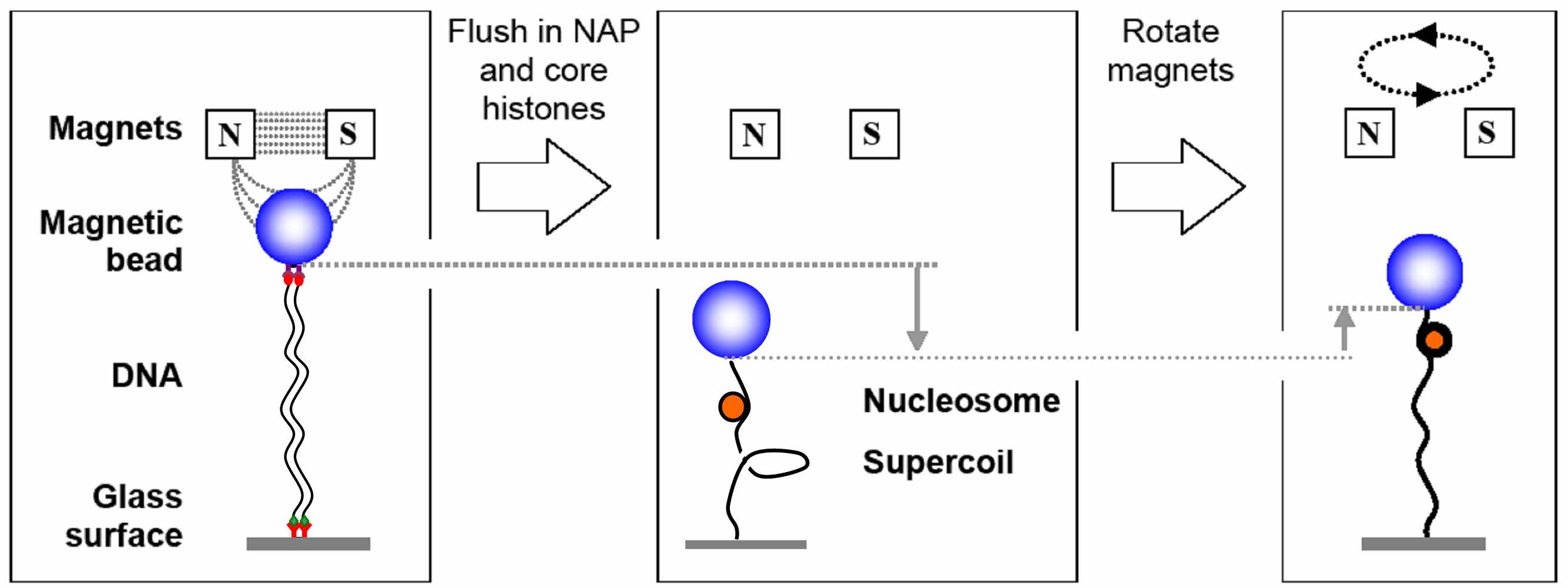
Assembly of nucleosomes mediated by the NAP1 chaperone, measured in the magnetic tweezers.