Student projects
We welcome students from a variety of backgrounds to participate in our research. Our group currently has the following opportunities for carrying out a bachelors or masters research project.
1. Building an artificial cell capable of growth and division
To understand the working principles of a cell, a powerful strategy is to build cellular functions step-by-step using well-defined components. This offers the additional advantage that such engineering strategies can potentially design products with enhanced properties in a controlled manner. Among which, two extremely important aspects include cellular growth, the step that preceded division, and abscission, both of which are required for proliferation and evolution.
The goal of this project is the reconstitution of cell growth, division and proliferation of synthetic cells. For this, the student will use state-of-art microscopy techniques to unravel the molecular mechanisms of cell growth and division in a minimalistic cell. We will reconstitute the minimum components to assemble functional cell-like compartments that will undergo repeated cycles of growth-division in a controllable way. The experiments will pave the way to understand the complex mechanical network at the fundamental biophysical level, and will help designing artificial cells with enhanced functional properties.
What we offer: We offer a solid well-structured facility for sample preparation and manipulation and state-of-art imaging, including multicolor confocal microscopy, fluorescence lifetime imaging microscopy and fluorescence correlation spectroscopy. Furthermore, the student will be closely supervised by an experienced team who will assist throughout the project. The student will also gain experience in project design, sample preparation, data acquisition, processing and presentation.
For further information, get in touch with Rafael Lira, even if informally: r.lira@[REMOVE THIS]tudelft.nl
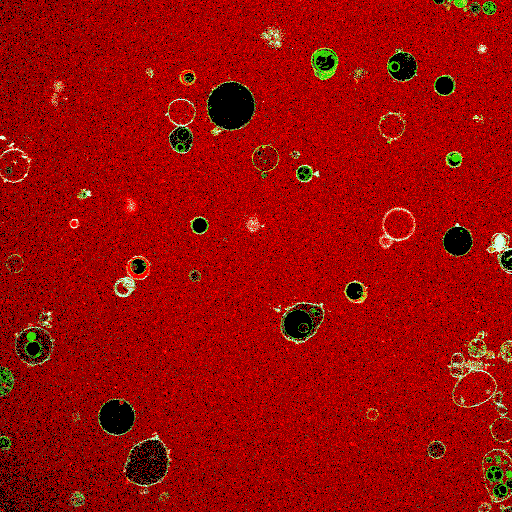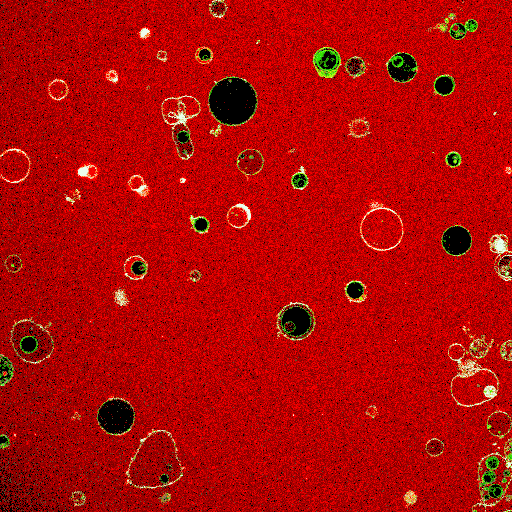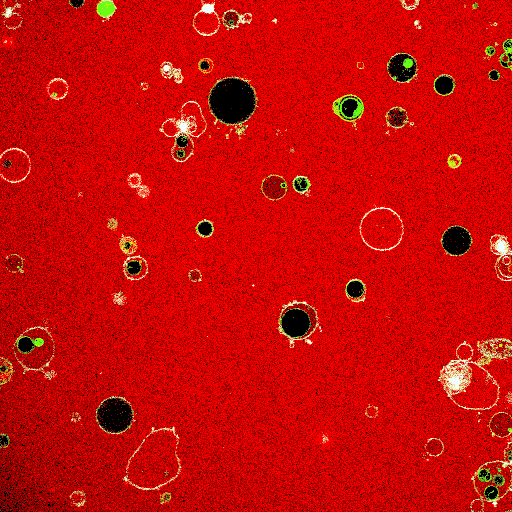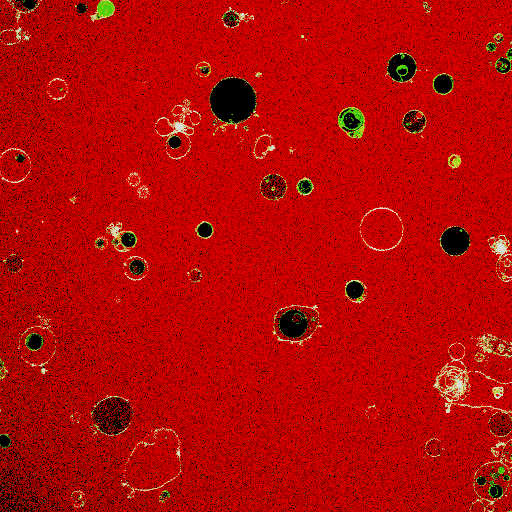
Synthetic cell-like lipid vesicles upon fusion of small liposomes. Fusion leads to increase in vesicle size, resulting in membrane fluctuations. The increase in compartment area is later used to aid division.
2. Building a living cell from scratch using microfluidics
Throughout the course of evolution life has developed a staggering complexity at the cellular level. To shed light on the fundamental blueprint of a cell and get a better understanding of the governing principles of cellular life, we are aiming to build a synthetic cell from the bottom-up using molecular building blocks (www.basyc.nl).
More specifically, we developed a microfluidic technology, Octanol-assisted Liposome Assembly (OLA), to produce cell-sized (5–20 µm) liposomes in our lab. These liposomes can be immobilized using microfluidic traps for further manipulations. In the past, we already succeeded in mimicking the form of rod-shaped bacteria by squeezing the liposomes into narrow confinements. Further, liposome growth was established by recruiting lipids from the external environment, and liposome division was induced by colliding them against well-defined microfluidic structures. Now the time has come to combine these modules into an integrated lab-on-a-chip system to establish a dynamic cycle of growing and dividing liposomes, mimicking a continuous life cycle of a living cell.
In this experimental and multidisciplinary project, you will gain experience working in a wetlab, learn how to operate a microfluidic setup, and perform basic light- and epifluorescence microscopy experiments. Based on these experiments we can further optimize parameters that are essential for the on-chip production of synthetic cells. If you are interested in this topic, please contact Bert Van Herck (B.VanHerck@[REMOVE THIS]tudelft.nl) for more information. Note that previous knowledge in biology or chemistry is helpful, but not necessary.
The controlled bubble-blowing process of Octanol-assisted Liposome Assembly (OLA), where monodispersed double-emulsion droplets are pinched off at the production junction to ultimately form unilamellar cell-sized liposomes.
3. Protein sequencing with nanopores
Nanopore technologies have been used by astronauts aboard the International Space Station and biologists travelling across the far reaches of the Earth to study DNA at the single-molecule level. However, analyzing the protein composition of cells at the single-molecule level, while an extremely valuable diagnostic tool, remains a difficult task. In this project we are taking important steps in developing nanopore technologies for proteomics.
Currently, we are exploring methods inspired by nanopore DNA sequencing. This approach involves attaching a small piece of protein (peptide) to a DNA strand to form a peptide-oligo conjugate (POC) which is pulled through a nanopore by a DNA motor enzyme. By measuring the ion current through the pore as the peptide moves through it, we can distinguish individual amino acids and detect important post-translational modifications! Our group has pioneered the proof-of-concept of this method, but engineering and data analysis breakthroughs are needed to push this technology to the next stage!
There is much to learn from this project, as it incorporates a multidisciplinary understanding of biology, biophysics, and biochemistry. You will gain experience working in a wet lab, learn how to handle datasets, and be involved in a very social lab environment. If you are a BEP/MEP/international student and interested to take part in this project, please contact Justas: j.ritmejeris[REMOVETHIS]tudelft.nl
We look forward to hearing from you!
4. Genome-in-a-Box: Bottom-up Reconstitution of Chromosome Organization
The molecular program of every cell is encoded in its genome. The genome size (the total number of DNA bases), varies over several orders of magnitude across organisms [1] (read more here). Despite this variety, all cells face the same critical challenge: to compact their genomes into the confined volume of a cell is a daunting task! Human cells have to compact about 1 m of DNA to 10 µm sized nucleus and bacteria about 1.4 mm of DNA to their micron sized nucleoids. How do cells do that? How do nanoscale proteins organize DNA into structures much larger then themselves? How does this influence the biophysical properties of DNA?
To contribute answers to these questions, we pursue the Genome-in-a-box project, where we isolate megabase-long DNA from live bacterium and combine it with purified proteins in microfluidic compartments which we then probe with fluorescent microscopy tools. This unique approach allows us to study the biophysics of large-scale chromosome organization, by analyzing the effects of individual DNA-organizing elements.
In context of this pursuit we have two open projects for either MEP or BEP students:
- Characterization of size and dynamical properties of microfluidically confined chromosomes (including radius of gyration, mesh size, coefficient of diffusion, motion correlations and deformability) [2] (read more here).
- Study of interaction of Structural Maintenance of Chromosome (SMC) proteins with isolated chromosomes [3] (read more here).
Both projects are experimental and will allow you to learn more about chromosome organization, DNA-protein interactions and microfluidics. Previous lab experience is helpful but not required. Do you find this interesting and want to learn more? Great! Please reach out to Martin Holub (m.holub[REMOVETHIS]tudelft.nl) or Alex Joesaar (a.h.joesaar[REMOVETHIS]tudelft.nl) and include a brief statement of motivation and description of your previous research experience in the email!

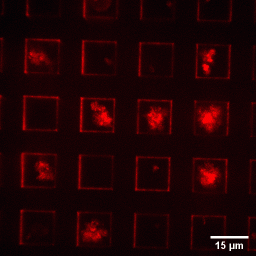
5. AFM-Fluorescence characterization of the plasmid partition protein ParB
A plasmid is a small, up to 2×105 pairs of bases, circular DNA molecule found inside various living organisms. The genes codified in these molecules are not essential as those codified in chromosomes, but provide some benefit for the cell (e.g. antibiotic resistance), hence, the importance of the cellular mechanisms responsible for the maintenance and organization of these molecules and their role in evolution.
