Restriction Enzymes
Biological function of type I restriction enzymes
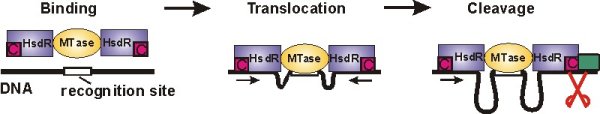
Type I restriction-modification enzymes provide bacteria with an efficient defense against viral DNA by cleaving it at random sites. They consist of a methyltransferase core unit (MTase), which can bind up to two restriction/motor subunits (HsdR). If the DNA substrate is hemimethylated at the recognition site DNA is recognized as own, bacterial DNA and methylation of the other strand follows. If the recognition site is not methylated it is considered hostile and DNA cleavage is initiated (see Figure below). Then the motor subunits start to translocate the adjacent DNA, thus pulling it towards the enzyme. This results in the formation of large DNA loops. Cleavage of the DNA finally occurs, when the translocation of the enzyme is blocked, e.g. by another enzyme, which is bound to the DNA.
We use single-molecule techniques like Atomic Force Microscopy (AFM) and magnetic tweezers to study all stages of the restriction reaction. The motor subunits of Type I restriction enzymes belong to the Superfamily 2 (SF2) of helicases. Therefore, beyond the particular interest for the Type I systems, this research is set into the broader context of understanding the mechanism and function of SF2 helicases. This group comprises such important enzymes as chromatin remodeling factors and the DNA repair enzyme Rad54. In particular, the ability of Type I restriction enzymes to introduce an enormous amount of supercoiling (see below) might be a shared property for some of the SF2 members.
AFM imaging of single translocating complexes
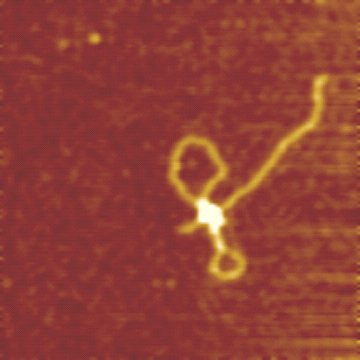
The enzyme-DNA complex can be imaged using AFM. It allows to resolve the different stages of the restriction reaction: binding of the MTase and the motor subunits, grabbing of the DNA by the motors, d.h.formation of the initial DNA loop, and the enzyme translocation. (see van Noort et al.)
The AFM image on the right shows a translocating EcoR124I complex on DNA (2364 bp DNA molecule with a single recognition site, 6 mM ATP, Scan range 500 nm). DNA translocation by EcoR124I results in the formation of the two visible DNA loops.
Magnetic tweezers investigations
Magnetic tweezers are an excellent tool to study the translocation of single enzyme complexes.
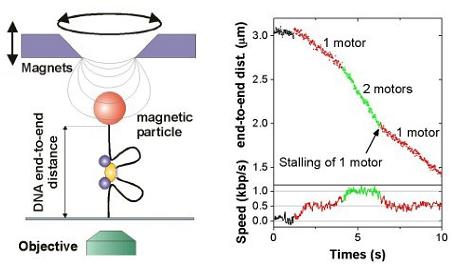
Magnetic tweezers allow to stretch and twist a single DNA molecule and to measure its end-to-end distance in real-time, while it is getting processed by an enzyme.
As Type I restriction enzymes pull in DNA, the effective DNA end-to-end distance decreases, which can thus be easily measured (see left image on the right). Applying the Type I restriction enzyme EcoR124I into the tweezers setup leads to characteristic time traces, where the DNA end-to-end distance is shrinking with constant velocity. The occurrence of two different velocities can be attributed to one ore two motor subunits being active in pulling DNA (see right image on the right). At room temperature a single motor unit can translocate the DNA with up to 550 bp/s. (For details see Seidel et al. 2004