Protein dynamics with nanopores
Protein movements are vital for every living cell, but hard to detect. Nanopore detection offers the broad time bandwidth needed to capture this fascinating nanoscale gymnastics. In this project we use DNA origami and nanopores to develop a novel a single-molecule detector that is well suited to study the broad time range of protein motions.
Proteins power our lives. Yet, despite their importance, major functional aspects remain elusive because of a lack of adequate analysis tools. Electrical detection using nanopores provides several advantages, one being the extensive time bandwidth from microseconds to hours. We exploit this to tackle the broad-range kinetics of proteins using a combination of solid-state nanopores with DNA origami.
Specifically, we anchor the protein of interest inside a nanopore and monitor its behavior by means of conductance changes over time (see Figure). This represents a radically new strategy to resolve the characteristic features of protein function, namely conformational changes, as well as the binding, processing, and release of substrates & cofactors. Currently, we investigate diverse systems, label-free: DNA processing, ligand binding specificity, etc. We observe remarkably detailed state trajectories, revealing protein dynamics far beyond mere binding and dissociation.
In combination with existing 3D structures, and molecular dynamics simulations, our approach opens a new way to unravel kinetic effects that were previously overlooked due to narrow bandwidths. Such broad-range kinetic information is urgently needed to understand protein function at the (sub-)molecular level.
This is a collaboration with the Hendrik Dietz Lab (TU Munich).
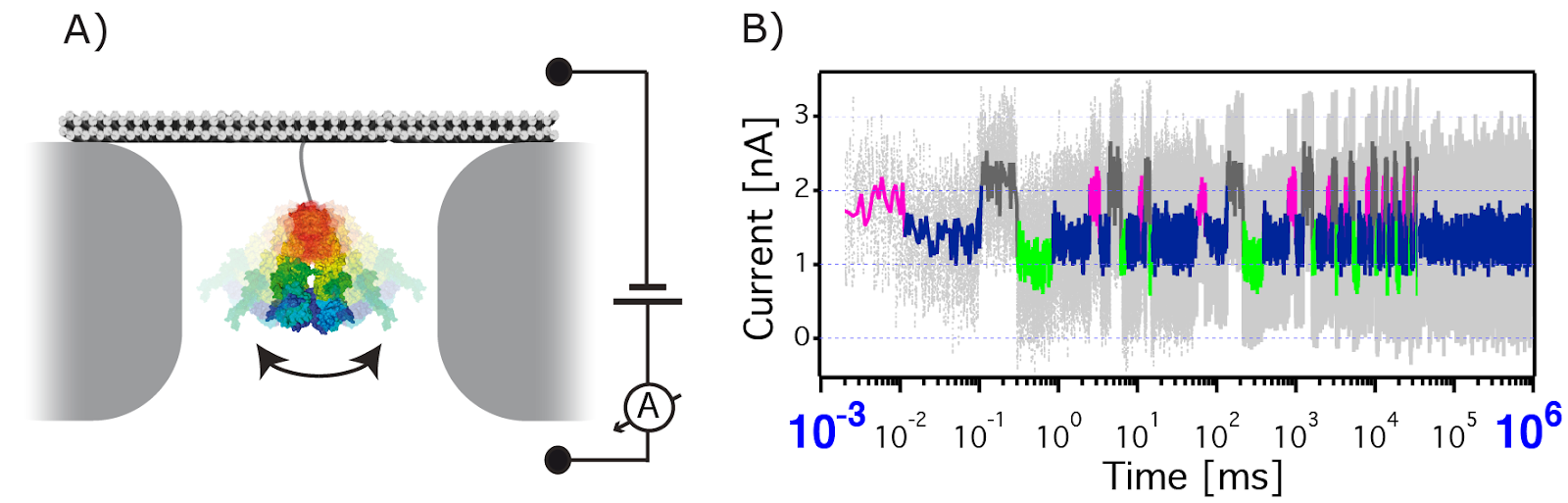
Detection of protein dynamics using solid-state nanopores. A) The target protein is localized inside a nanopore using a DNA-origami anchor. An electrical bias is applied across the pore causing an ionic current. B) High bandwidth current recordings show the modulations caused by protein conformational changes, as well as binding & processing of interaction partners. Four specific states were deduced by a Hidden Markov model from the filtered trajectory (colored accordingly). The simulated raw data is shown in gray.
People working on this project

Sonja Schmid

Allard Katan
- Room F0.190
- +31-(0)15-27 84561
- A.J.Katan@[REMOVE THIS]tudelft.nl
- Expertise: Scanning probe microscopy

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV


