Student projects
We welcome students from a variety of backgrounds to participate in our research. Our group currently has the following opportunities for carrying out a bachelors or masters research project.
1. Protein sequencing with nanopores
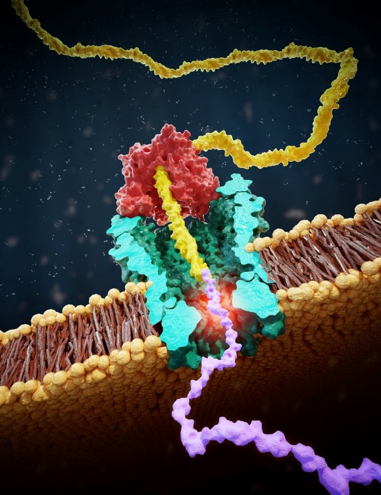
Protein sequencing at the single-molecule level, while extremely valuable for understanding biological processes and developing new diagnostic tools, remains a significant challenge. In this project, we are making important strides toward advancing nanopore technologies to study the protein composition.
Our sequencing approach is inspired by the DNA nanopore sequencing. This method involves creating peptide-oligo conjugates (POCs) by attaching a small protein fragment (peptide) to a DNA strand. A DNA motor enzyme then pulls this conjugate through a nanopore, where we measure changes in ion current to identify individual amino acids [1] and detect important post-translational modifications—small changes that can dramatically alter protein function [2, 3]. Our group has already demonstrated proof-of-concept for this method, but there is tremendous potential for molecular engineering and data analysis breakthroughs to refine and push this technology forward.
This project offers a multidisciplinary experience at the intersection of biology, biophysics, biochemistry, and data science. You can contribute to wet lab experiments and data analysis (Python, MATLAB), related to nanopore sequencing. Whether you’re interested in working in the lab or excited about coding, there’s a place for you in this project.
We’re looking for BEP/MEP/international students who are eager to take part in developing the future of nanopore-based proteomics.
If you’re interested in joining this exciting project, please contact Justas at j.ritmejeris[REMOVETHIS]tudelft.nl or Xiuqi at X.Chen-13[REMOVETHIS]tudelft.nl.
We look forward to hearing from you!
2. Building a living cell from scratch using microfluidics
Throughout the course of evolution life has developed a staggering complexity at the cellular level. To shed light on the fundamental blueprint of a cell and get a better understanding of the governing principles of cellular life, we are aiming to build a synthetic cell from the bottom-up using molecular building blocks (www.basyc.nl).
More specifically, we developed a microfluidic technology, Octanol-assisted Liposome Assembly (OLA), to produce cell-sized (5–20 µm) liposomes in our lab. These liposomes can be immobilized using microfluidic traps for further manipulations. In the past, we already succeeded in mimicking the form of rod-shaped bacteria by squeezing the liposomes into narrow confinements. Further, liposome growth was established by recruiting lipids from the external environment, and liposome division was induced by colliding them against well-defined microfluidic structures. Now the time has come to combine these modules into an integrated lab-on-a-chip system to establish a dynamic cycle of growing and dividing liposomes, mimicking a continuous life cycle of a living cell.
In this experimental and multidisciplinary project, you will gain experience working in a wetlab, learn how to operate a microfluidic setup, and perform basic light- and epifluorescence microscopy experiments. Based on these experiments we can further optimize parameters that are essential for the on-chip production of synthetic cells. If you are interested in this topic, please contact Bert Van Herck (B.VanHerck@[REMOVE THIS]tudelft.nl) for more information. Note that previous knowledge in biology or chemistry is helpful, but not necessary.
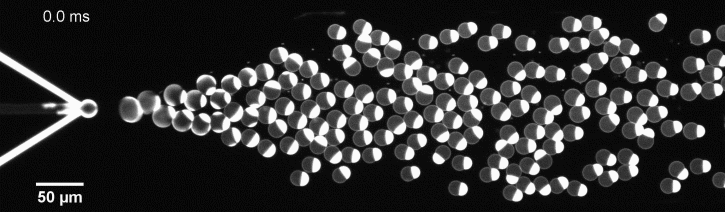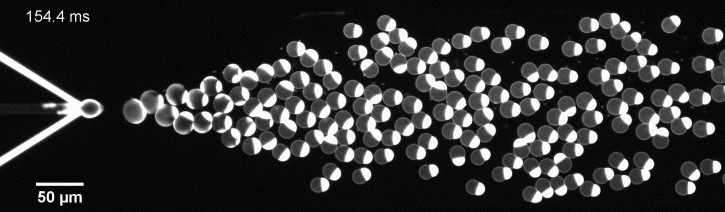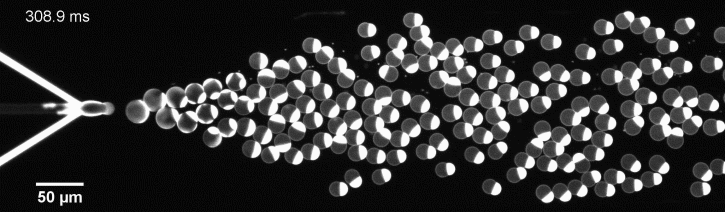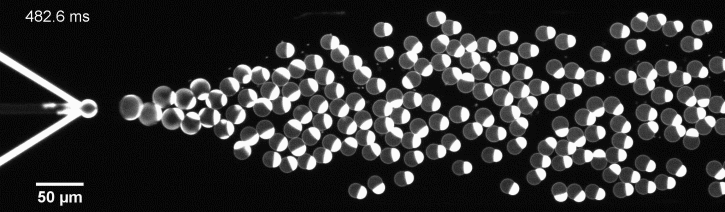
The controlled bubble-blowing process of Octanol-assisted Liposome Assembly (OLA), where monodispersed double-emulsion droplets are pinched off at the production junction to ultimately form unilamellar cell-sized liposomes.