Synthetic cell division
Bacterial division machinery
The idea of building a minimal artificial cell able to replicate itself has recently emerged as an ambitious goal that may be achievable in the long term. Following a bottom-up approach, we combine two elements required to mimic the fundamental functionality of cellular division: a protein machinery to carry out fission, and a compartment to enclose the chemical reactions defining the cell. As a minimal machinery we use FtsZ, a key protein indispensable for division in most bacteria. As confinements, we employ micron-size containers that are able to exchange energy with the environment, maintain the proteins functionality, and undergo morphological changes under the protein activity.
When encapsulated in liposomes, FtsZ assembles in filamentous bundles that can induce shrinkage of the vesicle. In liposomes with dimensions close to cell size, FtsZ can form a full ring. Realizing the importance of confinement dimensions on the protein spatial arrangement and activity, we developed a microfluidic technique to finely tune shape and size of these micron-size containers.
Also, to investigate the proper condition for FtsZ bundles to form, we employ water-in-oil droplets to study how relevant conditions, such as crowding and protein concentration, influence the condensation of FtsZ filaments into higher-ordered structures.
Furthermore, we observe that FtsZ strongly partitions on the surface of coacervates, liquid droplets generated by the spontaneous aggregation between polylysine and guanosine triphosphate, polymerizing into bundles and inducing a variety of fascinating morphological changes.
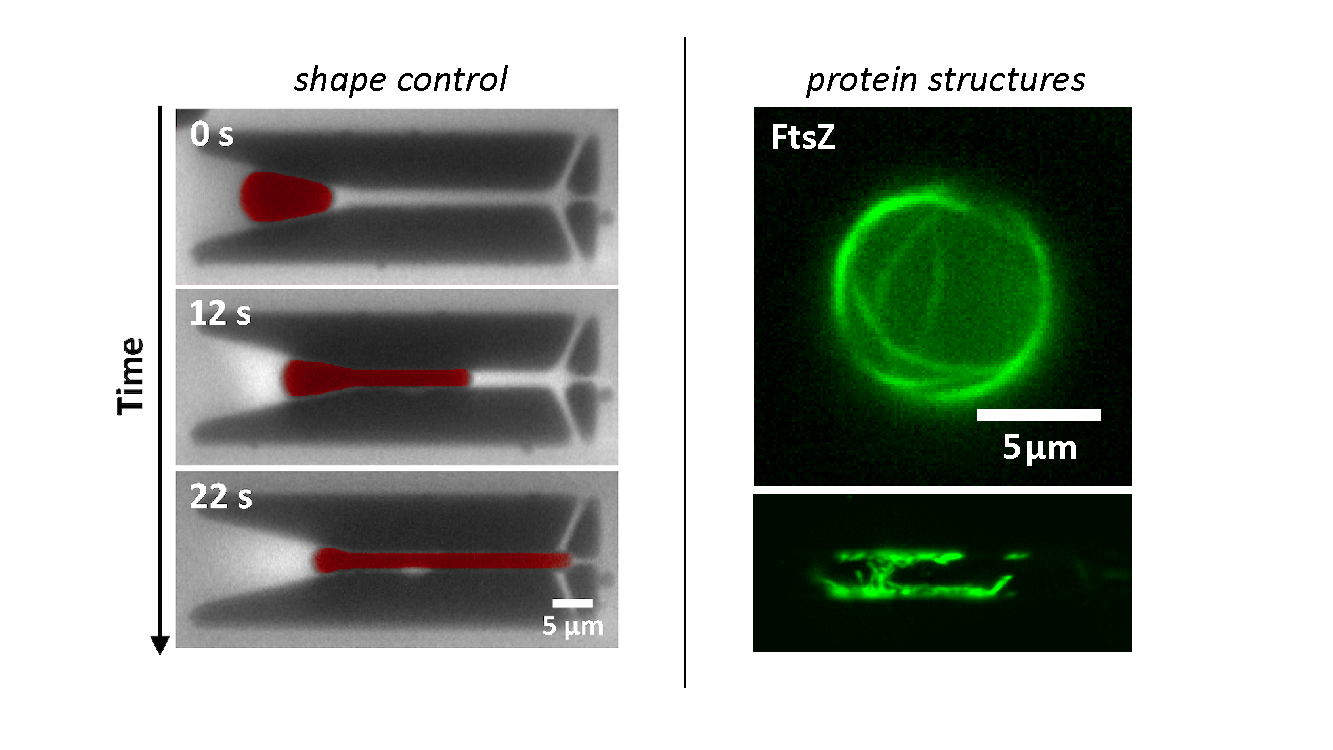
On the left: manipulation of artificial compartments into cell-like shape and size using microfluidics. On the right: reconstitution of FtsZ bundles inside water-in-oil droplets of different geometries (spherical and rod shape).
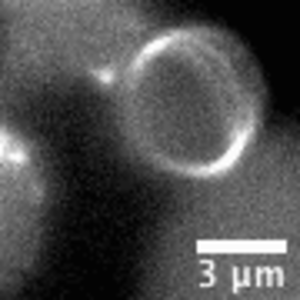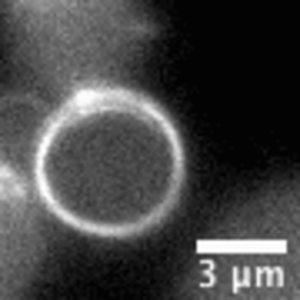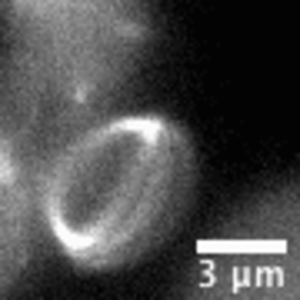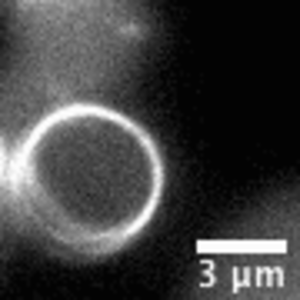
Video: In bacteria FtsZ is a key protein that assembles a constrictive ring localized at the mid cell, orchestrating the process of division. In the video, an FtsZ ring is artificially reconstructed inside a small unilamellar liposome.
Archeal division machinery
Life has many forms and, with them, many different ways of performing the same processes. Many archaeal species possess a unique and intriguing cell division machinery called the Cdv system. Composed of only 3 proteins, CdvA, B and C, it shares homology with many some components of the eukaryotic ESCRT machinery. However, its mechanism of action has been a long-standing enigma and it poses some very interesting questions about the evolutionary relation between eukaryotes and archaea.
We aim to reconstitute this whole system in vitro in a bottom-up manner. By using high resolution microscopy techniques such as AFM or TEM, we study the structure and arrangement of these proteins at the molecular scale. By understanding the interaction of these proteins with lipid vesicles, we aim to decipher how they act and drive the process of cell division, with the final aim of recreating a fully functional synthetic cell division machinery.
DNA-based cell division machinery
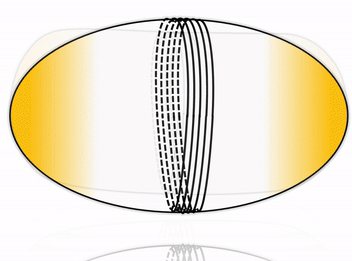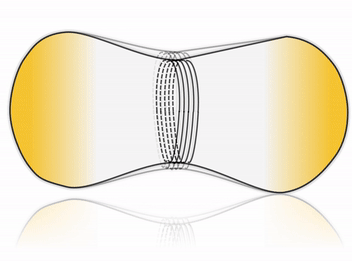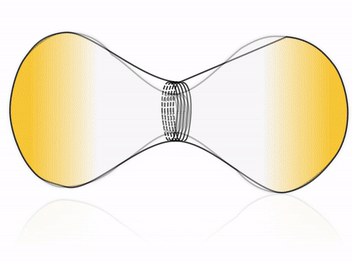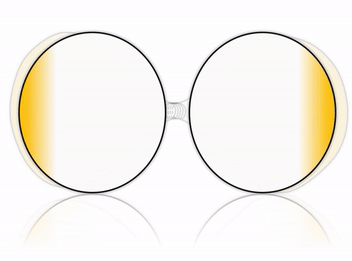
Cell division is one of the most fundamental features of life – perhaps even its defining characteristic. Many cells accomplish division by using a complex internal protein machinery operating with the same basic principle: a division ring assembles on the inner side of the cell membrane and constricts it until it pinches off the cell into two daughter cells. However, reconstructing the cell division machinery in vitro from its proteinaceous components still remains a formidable challenge, the main difficulty being to control the assembly and coordination of the division machinery in space and time.
To this goal, DNA nanotechnology offers unparalleled precision and flexibility in designing nanodevices. Impressive advance in this field has produced molecular nanomachines which can rival, and in some cases even surpass, the performance of their natural protein counterparts. We aim at harnessing the possibility of rational design offered by DNA nanotechnology to construct a fully synthetic, DNA-based division machinery in vitro. Inspired by cellular systems such as the ESCRT-III complex, we address this fascinating challenge by building an artificial division ring, able to assemble inside a liposome and undergo constriction, thereby driving the system towards fission.
People working on this project

Alberto Blanch Jover

Federico Fanalista

Nicola De Franceschi

Siddharth Deshpande

Jacob W.J. Kerssemakers
- Room F0.110
- +31-(0)15-27 88331
- J.W.J.Kerssemakers@[REMOVE THIS]tudelft.nl
- Expertise: Single-molecule instrumentation

Jaco van der Torre
- Room F0.190
- +31-(0)15-27 83959
- J.vanderTorre@[REMOVE THIS]tudelft.nl
- Expertise: Molecular biology

Eli van der Sluis
- Room F0.130
- +31-(0)15-27 86572
- E.O.vandersluis@[REMOVE THIS]tudelft.nl
- Expertise: Protein purification

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV







