Biomimetic nuclear pore complexes
Compartmentalization of DNA in the nucleus is one of the central features that distinguishes eukaryotic cells from less evolved organisms. Communication between nucleus and cytoplasm is achieved through a huge multiprotein complex, the nuclear pore complex (NPC), massive biological pores (~65 MDa) that act as gatekeepers of the nucleus. The central channel of NPCs is lined with intrinsically disordered proteins termed FG-Nucleoporins, or FG-Nups (rich in phenylalanine ‘F’ and glycine ‘G’ repeats), which impart a selective barrier. Small molecules (< 20-40 kDa) can freely diffuse through, whereas large cargoes (>40 kDa) are normally excluded, unless they are bound to a Nuclear Transport Receptor (NTR). In fact, NTRs can efficiently permeate through the FG-Nup barrier by hydrophobically interacting with their FG repeats, thereby lowering the energy barrier for translocation.
Despite the extensive literature on this subject, it is still unclear how NTRs manage to cross such a dense and cohesive meshwork, while still retaining a fast and efficient transport. Different models have tried to explain this intriguing process, yet a general consensus has not been so far reached. In our lab, we use nanopores to study the selective properties of minimalistic biomimetic NPCs, which are built by covalently tethering purified FG-Nups to the inner walls of a solid-state nanopore (Figure 1).
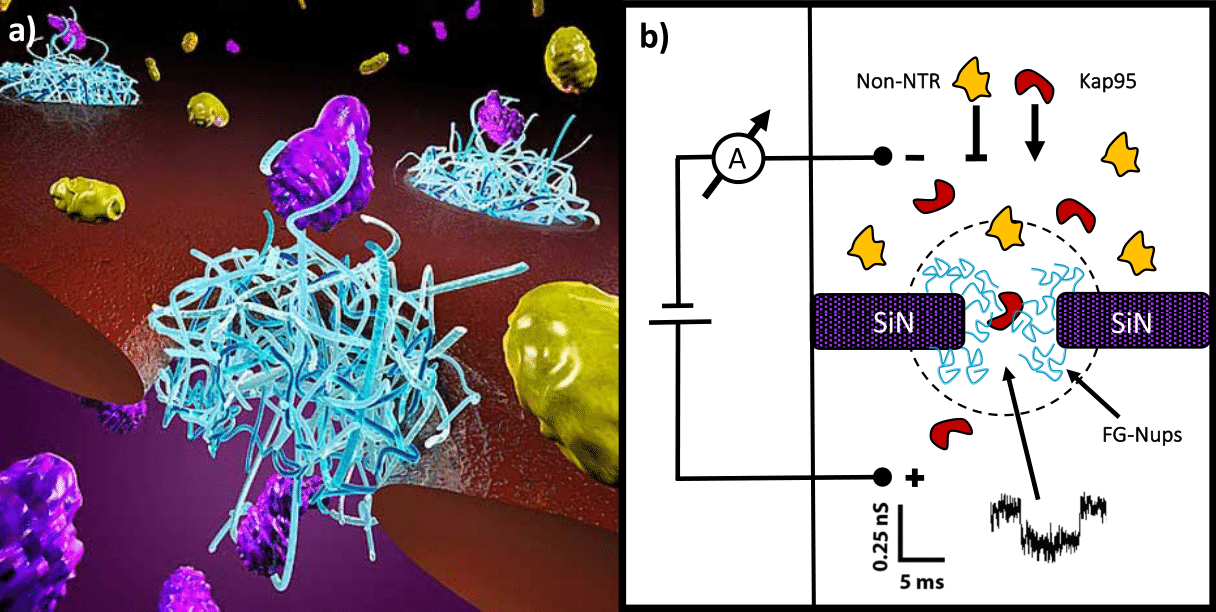
Figure 1. Biomimetic NPCs. a, Artistic representation of a Biomimetic NPC. b, Schematic of a solid-state nanopore functionalized with FG-Nups. Selectivity is tested by measuring the translocation efficiency of different cargoes (e.g. Kap95 or BSA) through the nanopore.
Mimicking protein import of the Nuclear Pore Complex with designer FG-Nups
In this project, we attempt at reconstituting NPC selective transport in biomimetic nanopores (Figure 2), using bottom-up designed artificial FG-Nups. By testing the impact of systematic variations of artificial FG-Nups on selectivity, we aim at unraveling what are the minimal features for having an efficient and selective transport. We believe this approach will finally shed light on some fundamental physical principles that govern the NPC behavior.
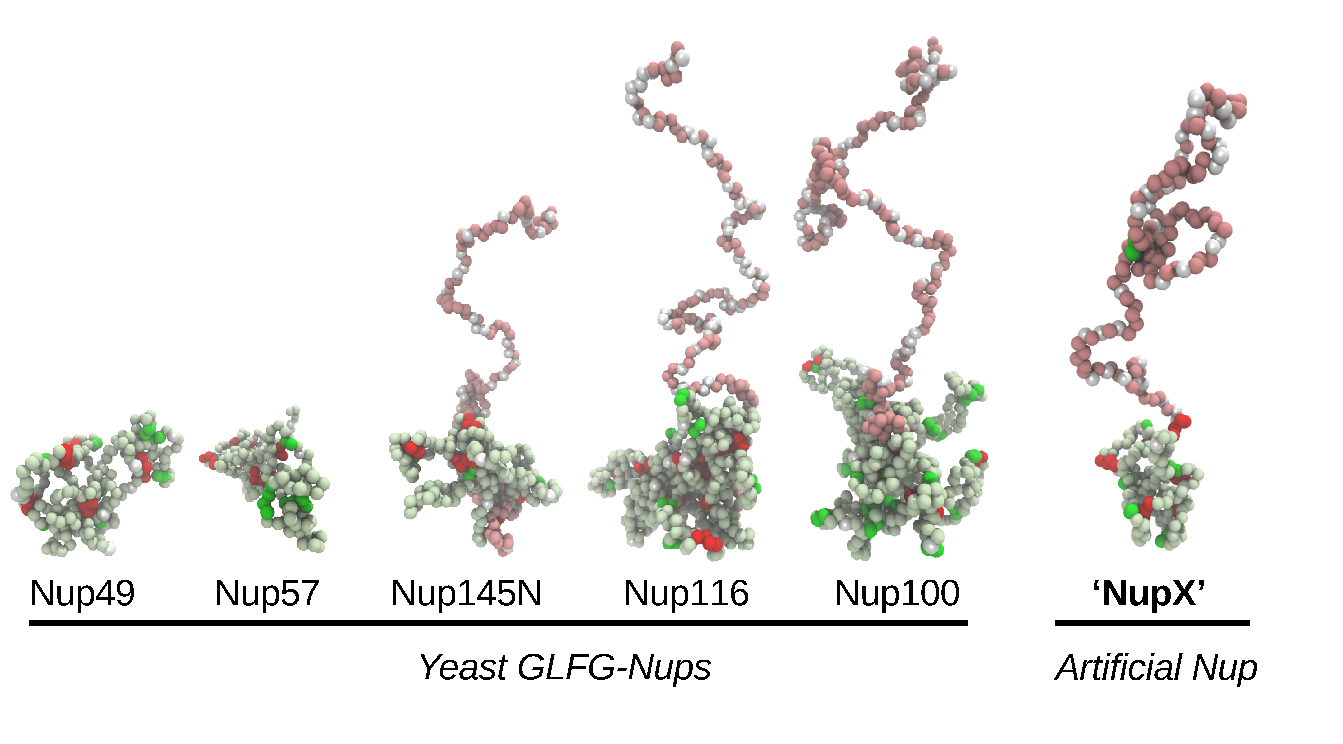
Figure 2. Simulation snapshots of isolated native yeast GLFG-type Nups. The conformations of Nup145N, Nup116, Nup100 highlight the so-called bimodality of the Nups, consisting of collapsed (light green) and extended (pink) domains. FG-repeats, GLFG-repeats and charged residues are displayed in green, red and white, respectively. The designed NupX adopts the same kind of conformations as GLFG-Nups.
DNA-origami scaffold for NPC mimics
With the help of DNA-origami technology (collaboration with the Dietz lab at Munich), we built a minimalistic version of the NPC, where FG-Nups are anchored to the inner walls of a ~30 nm large DNA-origami pore, with predefined stoichiometry and positioning (Figure 3). We employ this platform to study the arrangement of FG-Nups within the pore volume using AFM and TEM imaging techniques.
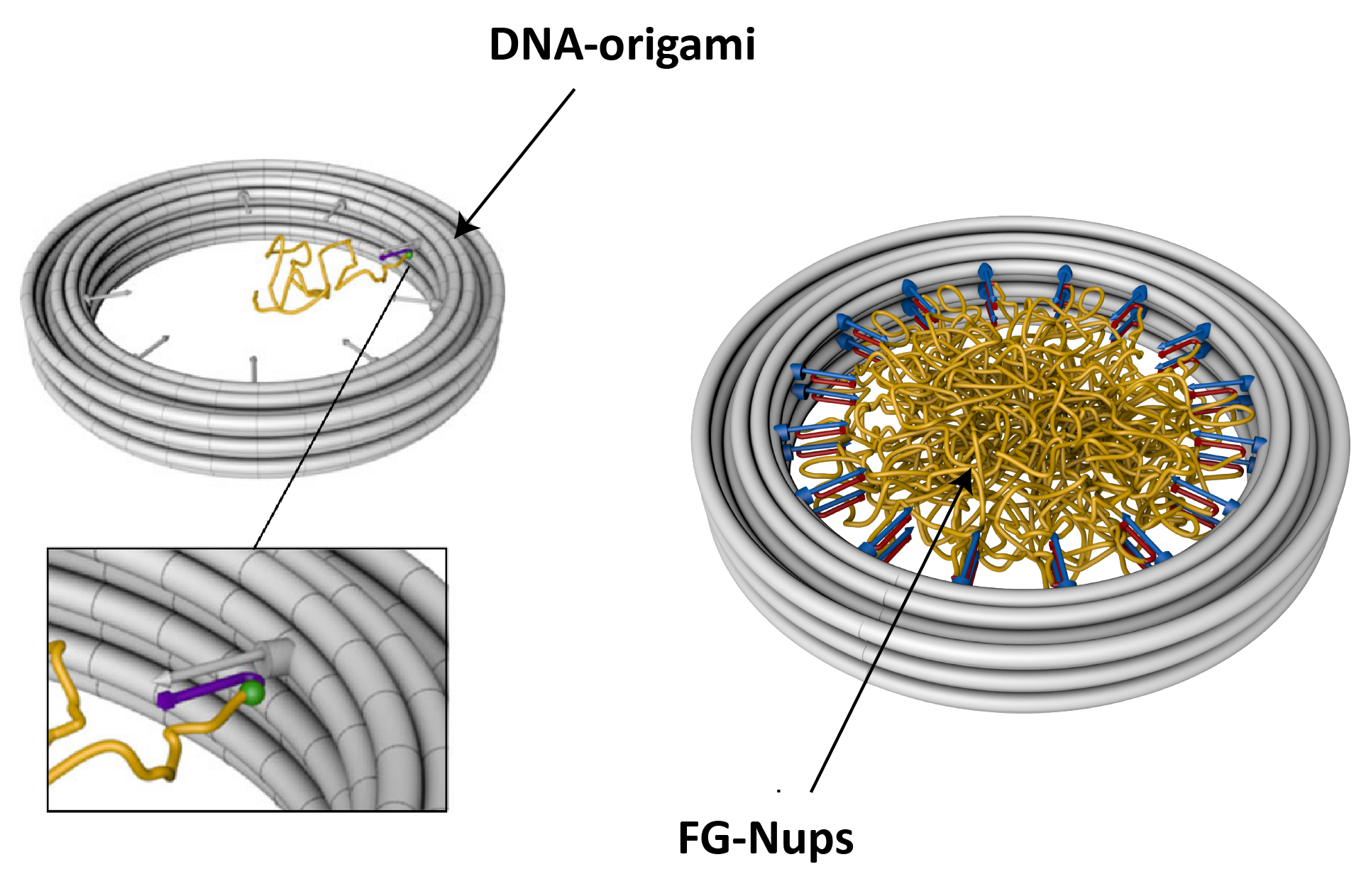
Figure 3. Schematics of DNA-origami ring with attached FG-Nups. FG-Nups are covalently attached to a single oligonucleotide, which in turn can hybridize to complementary oligo strands on the DNA-origami scaffold.
Mimicking mRNA export of the Nuclear Pore Complex
The goal of the mRNA export project is to recreate a minimal nuclear export system that is able to drive transport of mRNPs in a biologically relevant mimicking of NPC (Fig.4). Reducing the complexity of the yeast mRNA export system to its basic components in a bottom-up approach will allow us to unravel the nature of the minimal transport complex and describe its mechanism in details. To achieve this, we exploit the biomimetic nanopore technology. By coating solid-state nanopores with nucleoporins produced in-house, we use a current-base reading of single transport events of macromolecules through the biomimetic NPC.
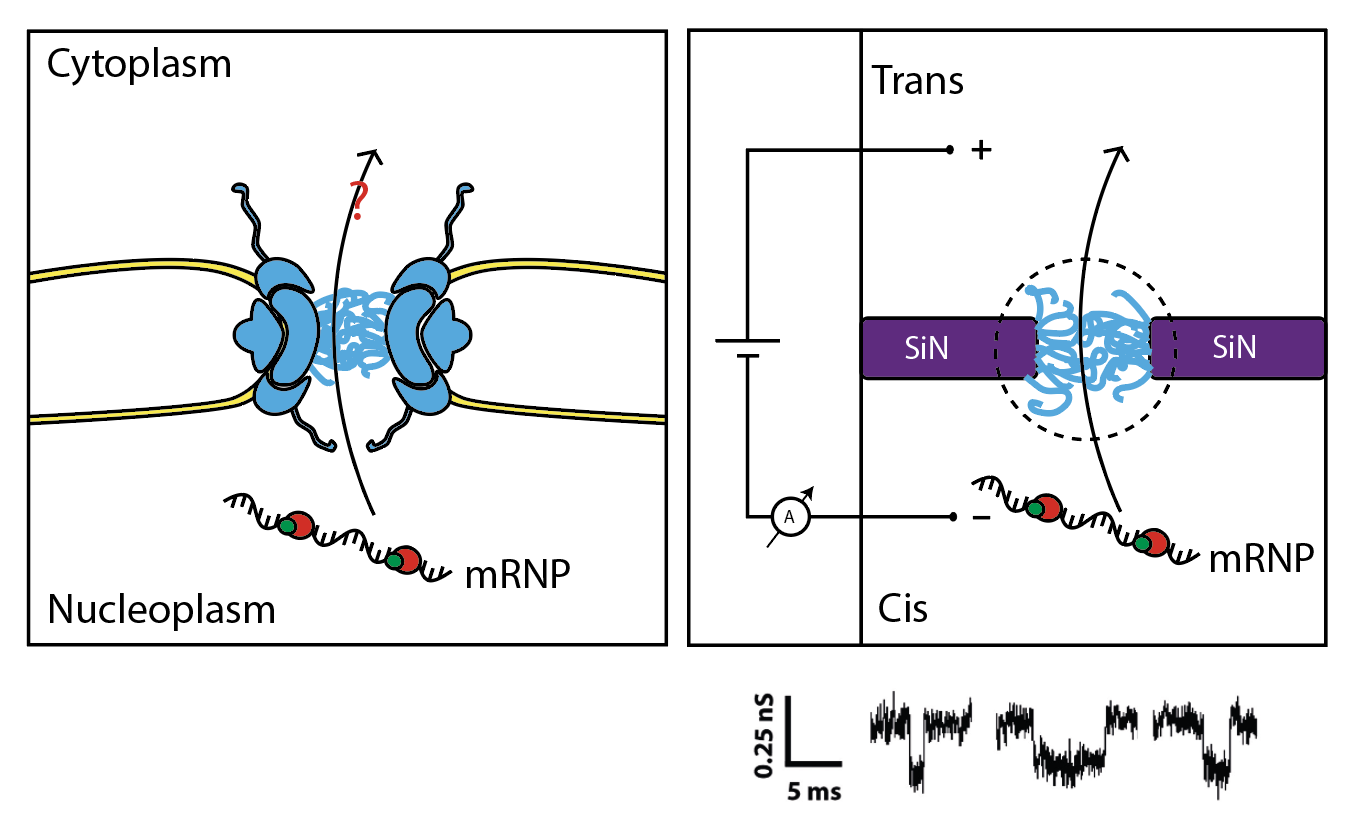
Figure 4. Illustration of mRNA export in cells (left) and in our biomimetic approach (right)
People working on this project

Alessio Fragasso

Paola De Magistris
Adithya Ananth

Eli van der Sluis
- Room F0.130
- +31-(0)15-27 86572
- E.O.vandersluis@[REMOVE THIS]tudelft.nl
- Expertise: Protein purification

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV



