Bacterial cell division
Cell division is a basic requirement for life, and in bacteria this process is a major target for antibiotics. Despite this, the mechanism of bacterial cell division remains poorly understood. During division, dozens of proteins dynamically interact with one another across large distances to do several tasks, such as (i) accurately separate chromosomes, (ii) place the division ring at the correct location, (iii) constrict the cell envelope inward against high turgor pressure, and (iv) separate from one another.
We have shown how the shape and size of cells affects the dynamic spatial organization of the Min protein oscillators in E. coli that are involved in division ring placement. In this project, we used nanofabrication and chemical treatment to sculpt cells into a variety of shapes and image Min dynamics, finding a wide variety of oscillation patterns governed by the cell boundary. (see our paper F. Wu et al, Nature Nanotechn.,
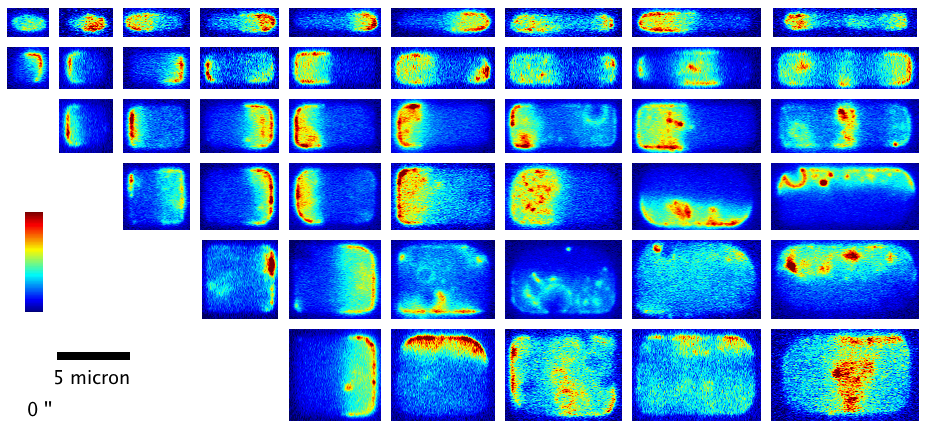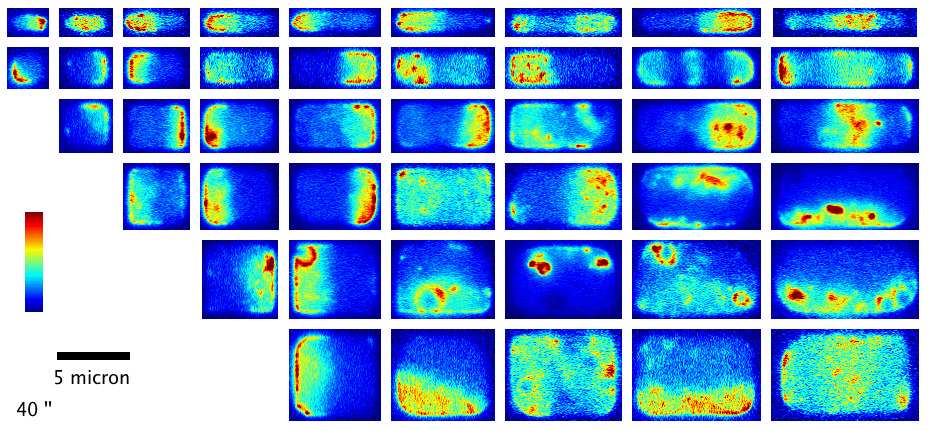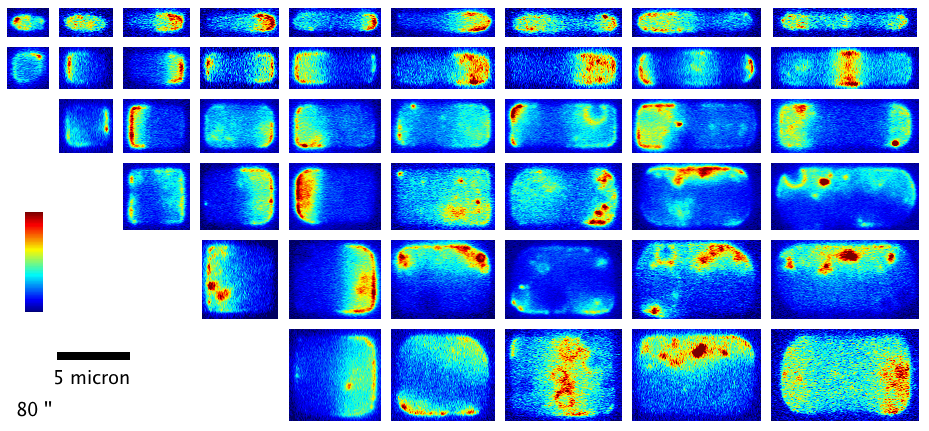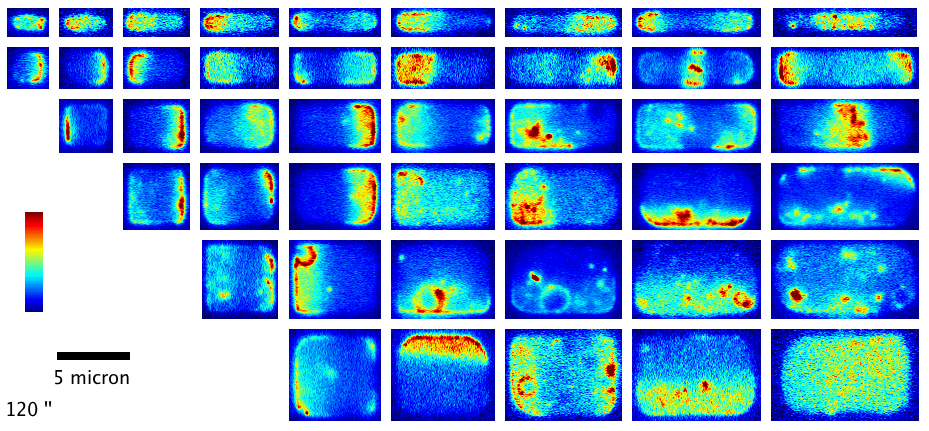
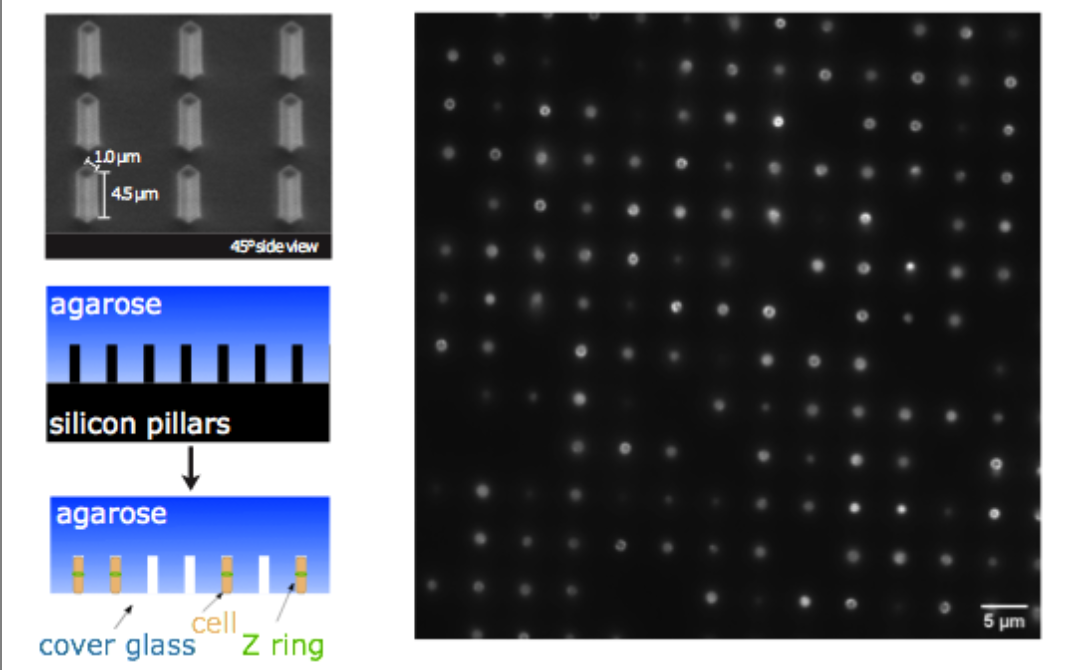
More recently, we have investigated the dynamics of the cytoskeletal protein FtsZ in live B. subtilis cells, in collaboration with the Holden lab at Newcastle University (UK). In this project, we created ‘microholes’ that can be used to confine bacteria vertically so that the full division ring can be observed in a single imaging plane. Our groups found that FtsZ forms motile filaments in actively constricting division rings rather than static structures as previously thought. Currently, we are expanding on this work investigating how cytoskeletal dynamics are coordinated with the activities of other essential division proteins, especially the enzymes that synthesize cell wall material. We do this by combining the ‘microhole’ technology with microfluidics so that we can instantly perturb individual components of the division machinery with drugs while imaging their dynamics.
People working on this project

Kevin Whitley
Fabai Wu

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV

