Bacterial chromosome organization
Organisation of the circular chromosome in E.coli
Although the physical properties of chromosomes, including their morphology, mechanics, and dynamics are crucial for their biological function, many basic questions remain unresolved.
In this project, we directly image the circular chromosome in live E. coli with a broadened cell shape. We find that it exhibits a torus topology with, on average, a lower-density origin of replication and an ultrathin flexible string of DNA at the terminus of replication. At the single-cell level, the torus is strikingly heterogeneous, with blob-like Mbp-size domains that undergo major dynamic rearrangements, splitting and merging at a minute timescale. Our findings provide an architectural basis for the understanding of the dynamic spatial organization of bacterial genomes in live cells.
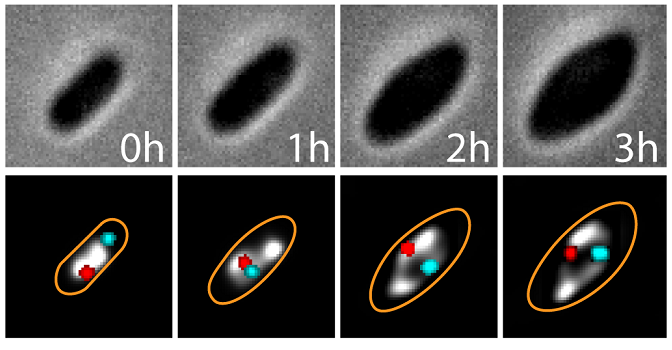
Fig.1 Widening the E.coli cell. The circular E. coli chromosome exhibits a toroidal donut shape that can be visualized upon cell expansion. Time-lapse fluorescence images of an E. coli cell growing into a lemon shape. Top panel, phase contrast image; bottom panel, overlay of Ori focus (red) and Ter focus (cyan) on a grey-scale deconvolved image of the chromosome labeled by HU-mYPet. Time is indicated in hours.
Cell Boundary Confinement Sets the Size and Position of
In this project, we show in Escherichia coli that spatial confinement plays a dominant role in determining both the chromosome size and it’s position. In E.coli cells with lengths up to 10 times the normal size, single chromosomes are observed to expand only ~4-fold in size. Chromosomes show pronounced internal dynamics but exhibit a robust positioning where single nucleoids reside robustly at mid-cell, whereas two nucleoids self-organize at 1/4 and 3/4 positions. The cell-size-dependent expansion of the nucleoid is only modestly influenced by deletions of nucleoid-associated proteins, whereas osmotic manipulation experiments reveal a prominent role of molecular crowding.
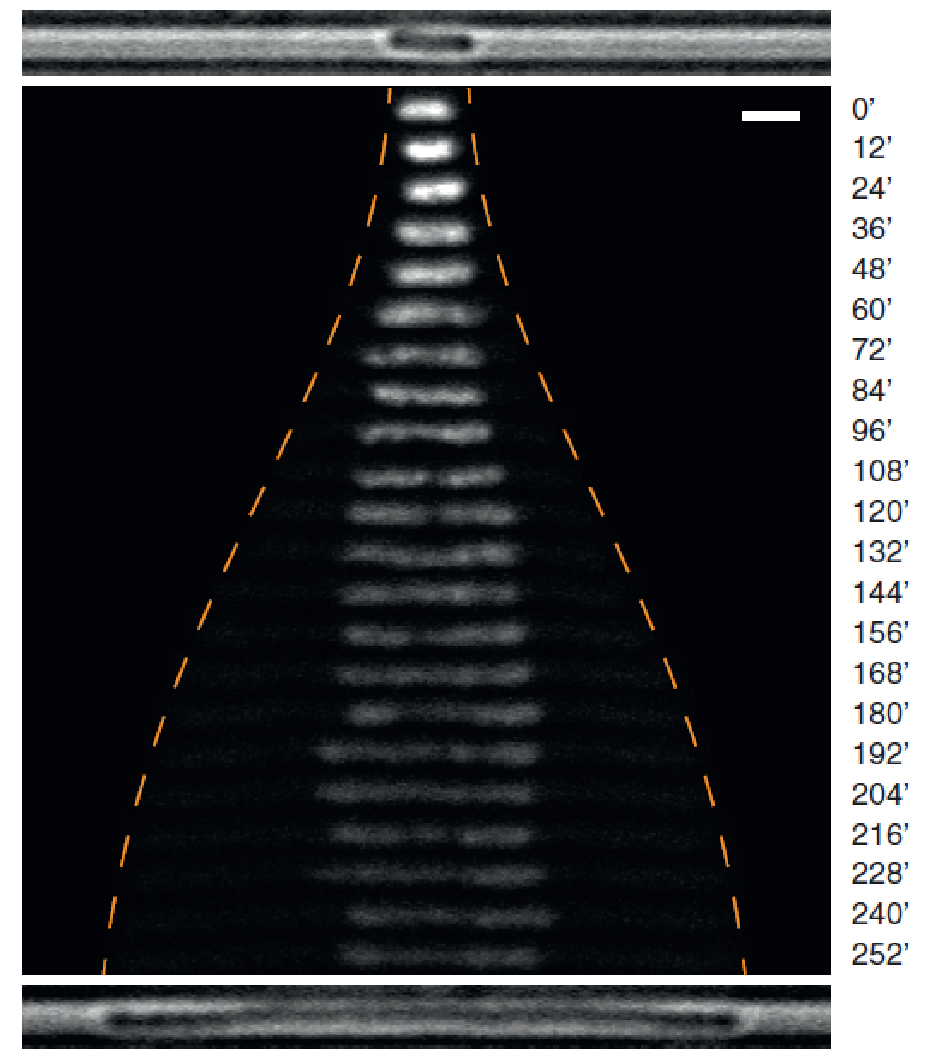
Fig.2 Chromosome Size and Positioning Are Dependent on Cell Size: Time-lapse images of a labeled chromosome as it expands with cell growth. The orange dashed line indicates the positions of the cell poles. Time is indicated in minutes.
Influence of antibiotics on bacterial chromosomes
Diseases caused by various bacteria constitute one of the biggest public health problems worldwide, causing ~15 million deaths per year. Therefore improving our understanding of the antibiotic function as well as bacterial resistance mechanism is of paramount importance.
In our lab, we developed a novel method to manipulate bacterial cell shapes and by doing so we investigate the finer structure of E.coli chromosome. In the current framework we plan study the influence of antibiotics on the chromosomes of live bacteria, by using fluorescence microscopy and quantitative data analysis (Fig.3).
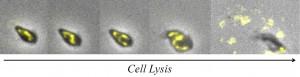
Fig3. Time-lapse images of an E.coli cell lysis. DNA inside the cell is labelled with fellow fluorescent protein.
People working on this project

Aleksandre Japaridze
- Room F0.170
- +31-(0)15-2788331
- A.Japaridze@[REMOVE THIS]tudelft.nl
- Research scientist startup company
Fabai Wu

Jaco van der Torre
- Room F0.190
- +31-(0)15-27 83959
- J.vanderTorre@[REMOVE THIS]tudelft.nl
- Expertise: Molecular biology

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV




