Optical nanopores
Solid-state nanopores are single-molecule sensors that hold great potential for the next generation of protein and nucleic-acid analysis. Despite their many opportunities, the conventional ionic current detection scheme that is at the heart of the sensor suffers inherent limitations. This scheme intrinsically couples signal strength to the driving voltage, requires the use of high-concentration electrolytes, may suffer from capacitive noise, and poses challenges to high-density sensor integration. Hence we explore how optical readout can facilitate nanopore detection of biomolecules, in various ways:
Optical Nanopores for label-free detection of biomolecules
In this project, we are aiming to address these limitations by integrating nano-optics with nanopore sensing. In virtue of the state-of-art nanofabrication techniques, we build nano-scale antennas and waveguides on the top of nanopores to manipulate the light-matter interaction at the single molecule level. Such combination enables the new readout approaches such as scattering, transmission and fluorescence; and control handles like optical forces, besides the typical ionic current and the electric field that nanopores provide. As a result, we are able to detect single biomolecules with plasmonic scattering or plasmon-enhanced transmission without the need of labels, and to trap and tweeze protein molecules with the confined and enhanced electromagnetic field. And probably in the near future, we can acquire molecular information such as their structures and conformations by the optical readout on nanopores.
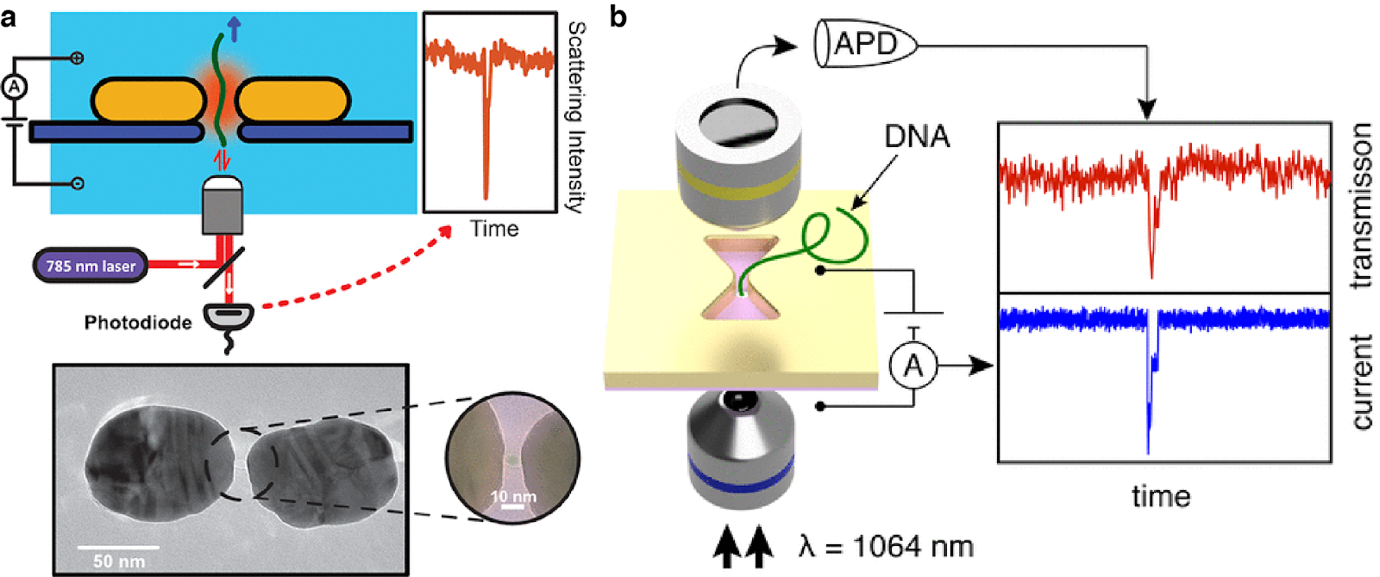
Figure 1. Label-free DNA detection using plasmonic nanopores.
Single fluorophore detection for nuclear pore selectivity and protein sequencing
Holes in metal films have the property to block light, if the light’s wavelength is larger than the diameter of the holes. These subwavelength apertures are also called Zero Mode Waveguides. Molecules, on the other hand, can still diffuse through the holes. We use this phenomenon to detect fluorophores after they have traveled through the metal membrane and thereby traversed from a dark region to an illuminated side. After passage, the fluorophores are excited and emit light, which can be detected. We use this property both for background suppression and in order to prevent bleaching before travelling through the aperture. After the passage they will quickly diffuse away and thus make it possible to detect single fluorophores. Currently we are working on the reliable detection of single freely diffusing fluorophores. We are working towards applying this technique to study two questions: Selectivity in nuclear pore transport and protein sequencing.
For the first project, the membrane separating the reservoirs represents the nuclear membrane and the nanoholes are modified to mimic the nuclear pore complex by grafting them with nucleoporin proteins. Similar measurements have been done by monitoring the current blockade in our lab already. These electrophysiological methods set requirements to the applied voltages and salt solutions, though. The optical detection of the analytes has the advantage that analytes can be distinguished from a mixture by their colour. Additionally it gives complete freedom for the choice of voltage and salt used. In addition the system’s noise does not depend on the modification of the pore.
In order to achieve protein sequencing, we are building on previous studies showing that it is enough to detect the sequence and distances of only two amino acids within a given protein of the human proteome to identify it with a high degree of certainty. Blockade of light by zero mode waveguides is crucially needed. We investigate mechanisms that sequentially feed proteins to the detection volume and thus translate a spatial distance to a temporal distance in the detection.
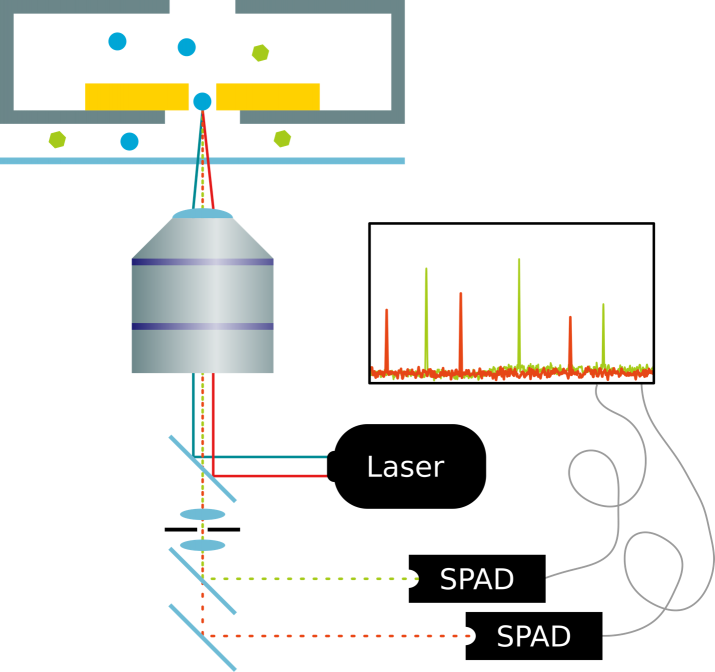
Figure 2: Single fluorophore detection in zero-mode-waveguide nanopores.
Optical trapping of single biomolecules
The ability to tweeze and interrogate label-free single proteins opens an interesting new avenue for the physical properties such as shape and size to be probed. We show label-free optical trapping of individual 20 nm beads and individual proteins such as Beta-Amylase, CRISPR/Cas9 using inverted bowtie shaped plasmonic gold nanopores. The plasmonic gold nanopores afford 2 key features. First, the transmission of light through these apertures are extremely sensitive to local changes in the refractive index. There is a shift in resonance when a molecule is trapped, allowing us to detect the molecule in a label-free optical manner. Second, the gold plasmonic aperture focuses light down to sub-diffraction spots thereby increasing the optical gradient. The enhancement of the optical gradient provides an optical trapping force that allows molecules to be trapped for upwards of seconds, opening up new routes to acquire information on the proteins’ shape, conformation and dynamics. On demand, we can release the trapped molecule by turning off the laser, allowing the molecule to diffuse back into the reservoir and for another protein to be interrogated. Current experiments investigating protein conformational changes are ongoing.
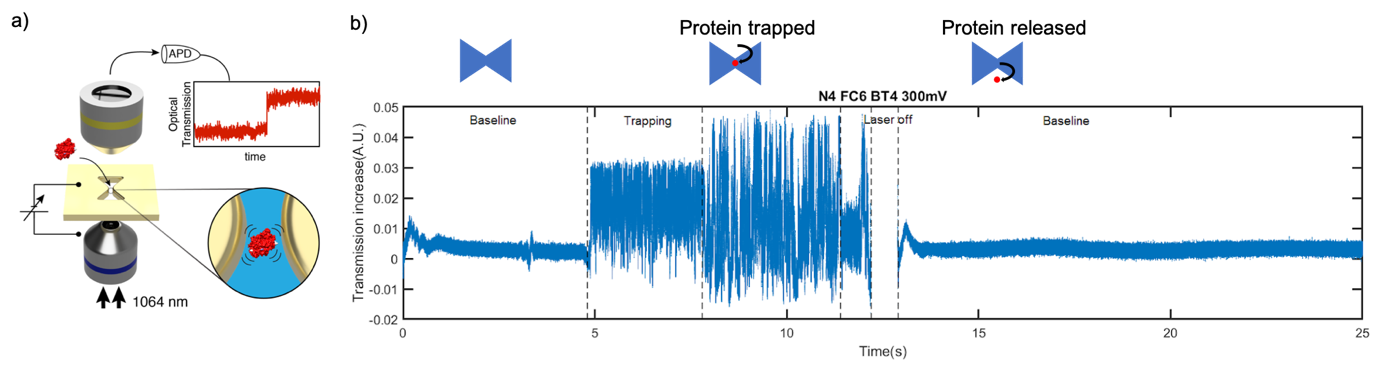
Figure 3. Tweezing of individual proteins in plasmonic nanopores. There is an increase in the transmission of the aperture when a protein is trapped. Different amplitudes in the increase of the transmission signal is observed, suggesting different conformational state. After the laser is turned off and back on, the signal returns to the same baseline level as the level before the start of the event, indicating the trapped protein was released.
People working on this project

Xin Shi

Nils Klughammer

Wayne Yang
Daniel Verschueren

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV



