Protein sequencing with nanopores
The protein composition of a cell or tissue is an ideal diagnostic indicator. We work towards single-molecule identification of proteins using nanopore technology.
Protein fingerprinting with nanopores
Proteins are major building blocks of life. Among them are crucial molecular machines – the actual makers in the cells. The protein content of a cell or organism provides therefore key information about biological processes and disease. Despite the importance of protein analysis, only a handful of techniques are available to determine protein sequences and these methods are expensive, time-consuming, and require huge & sophisticated instrumentation.
During the last few decades, nanopore technology has emerged as one of the most promising techniques to sequence DNA. Sequencing proteins at the single-molecule level is a clear next goal. This however presents even greater challenges than DNA sequencing, due to the overall higher complexity of proteins (20 different amino acids instead of 4 DNA basis, folded 3D structure, diverse charges, etc.).
Currently, we focus on protein fingerprinting: using protein nanopores, we work on protein identification rather than de novo sequencing. We pursue various approaches, e.g., we designed artificial detection tags that create recognizable protein fingerprints. Combining this approach with modern machine learning techniques and existing proteome databases, we work towards cheap & fast proteome quantification, with single-molecule resolution.
This is a collaboration with the Chirlmin Joo Lab (Delft), Giovanni Maglia Lab (Groningen), Rienk Eelkema Lab (Delft).
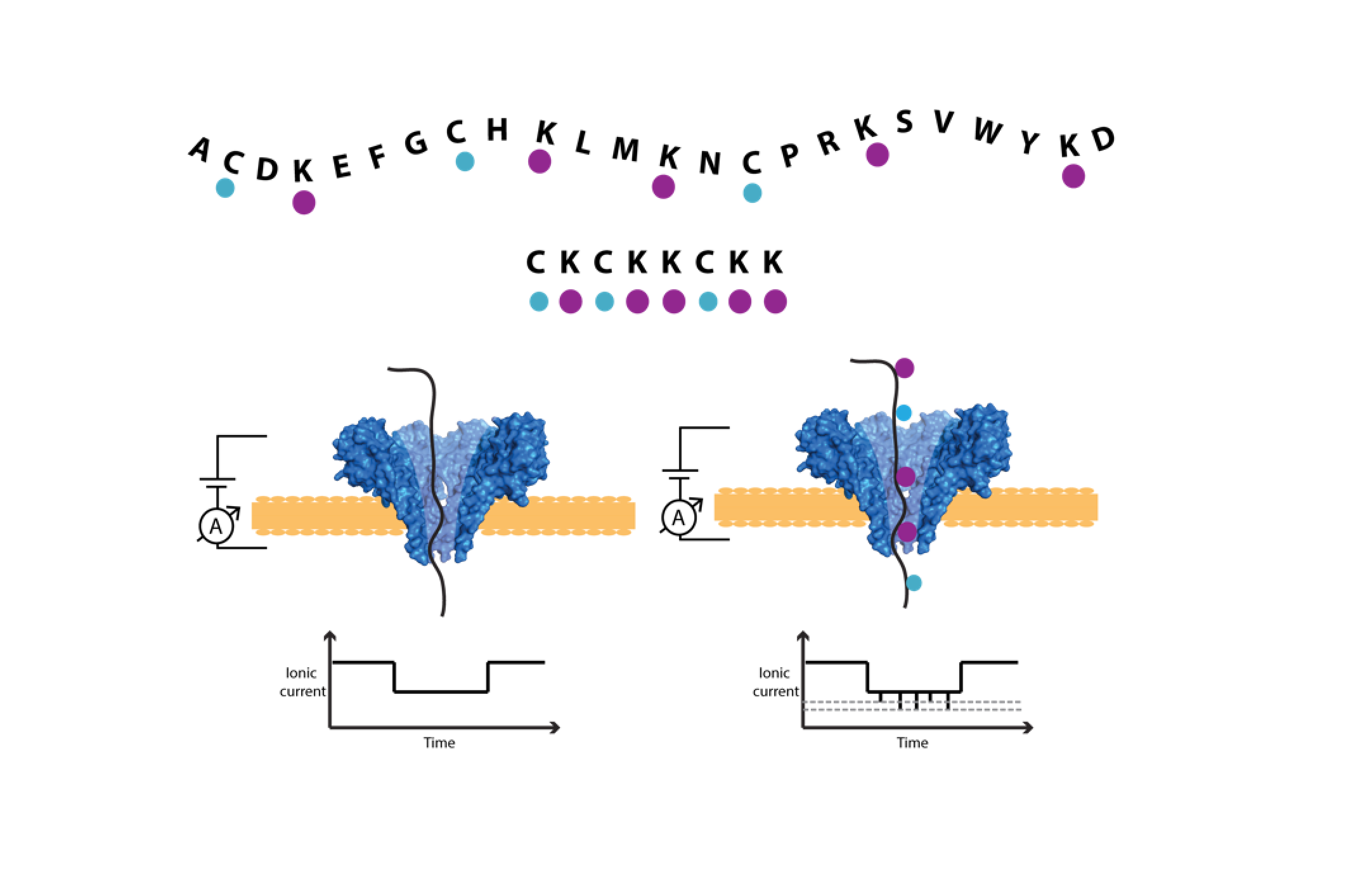
Protein sequencing with nanopores
Complementing the protein sequencing approaches using tagged amino acids, we are also investigating methods more directly inspired by nanopore DNA sequencing. Intrinsic to this approach is a method for slowing and controlling the translocation of the analyte through the nanopore. In nanopore DNA sequencing, this is accomplished by using a DNA translocase bound to the DNA, such as a helicase or polymerase, which is too big to fit through the nanopore. This bulky enzyme arrests the motion of the DNA through the pore, and as it walks, it moves the DNA through the pore step by step. This allows for a distinct measurement of the ion current through the nanopore at each DNA position, from which we can infer the sequence of bases. We are working on developing a similar scheme for controllably moving proteins through nanopores, and interpreting the resultant ion current signal to identify the protein primary structure.
People working on this project

Laura Restrepo Perez

Sonja Schmid

Henry Brinkerhoff

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV



