SMC proteins for DNA loop extrusion
In a human cell, approximately 2 meters of DNA are compacted into micrometer-sized chromosomes. When cells divide, DNA is distributed equally into daughter cells, such that each of them inherits exactly one copy of the genome. It remains a mystery how DNA is packaged within confines of cells and equally divided when cells divide. Recently, our group (in collaboration with Christian Haering, EMBL Heidelberg) elucidated the underlying mechanism of DNA organisation: a process called ‘loop extrusion’ that condenses DNA by progressively pulling DNA into many loops by the ring-shaped protein complex condensin, a a member of the SMC (structural maintenance of chromatin) protein complexes.
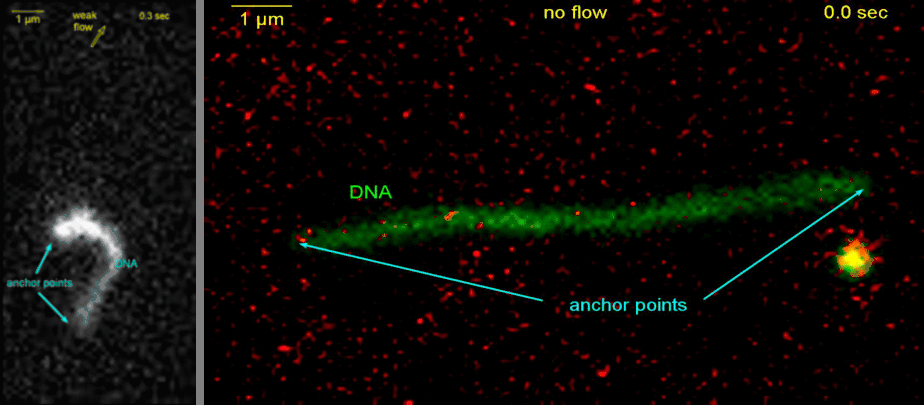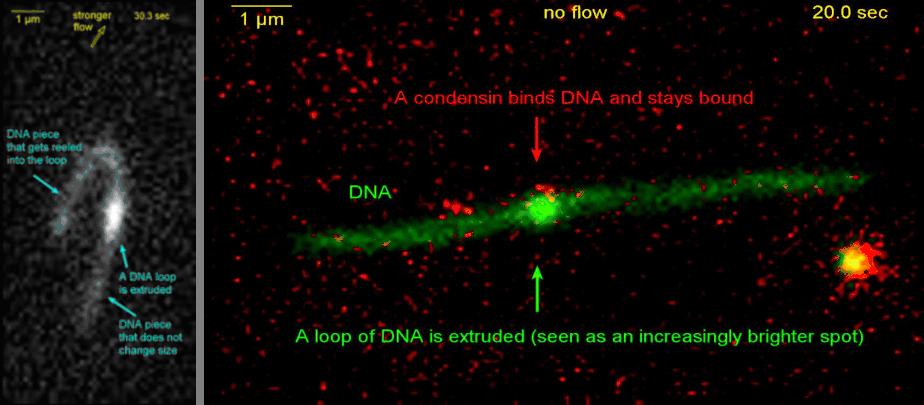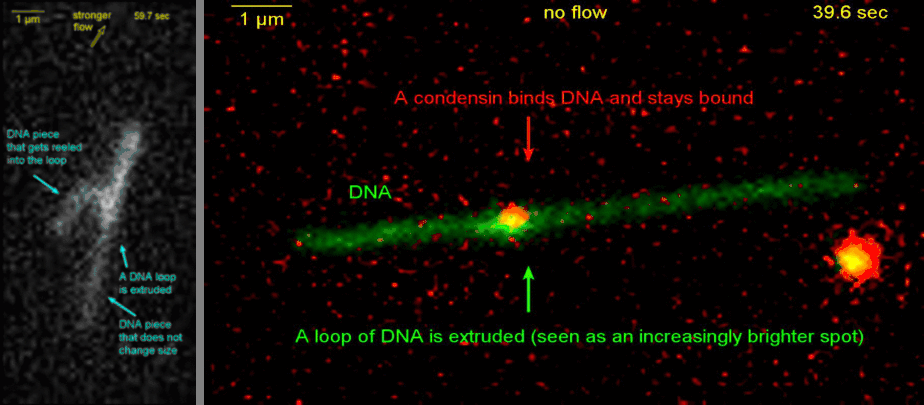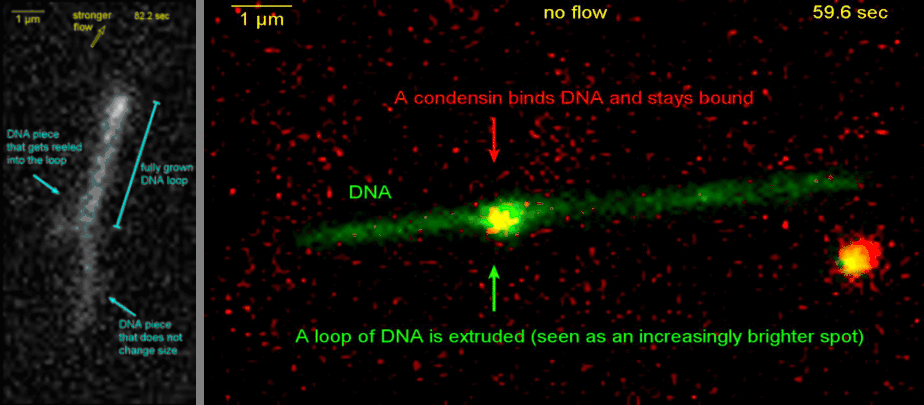
Loop extrusion by condensin: from bare DNA towards chromosome organization
Structural maintenance of chromosome (SMC) is a class of protein complexes that organizes DNA into the sister chromatids in the mitotic phase of a cell cycle. We employ various single-molecule assays like magnetic tweezers, fluorescence microscopy, and atomic force microscopy to elucidate the mechanical properties of SMC proteins. We recently showed that condensin compacts DNA upon ATP hydrolysis (Fig.1) and the underlying mechanism of DNA compaction is by extruding loops on DNA in a progressive manner. (Fig.2). The dynamic visualization of the loop formation flooded us with details like the velocity and step size of the condensin motor, asymmetric nature of the loop extrusion, and the size of the loops; which disclosed the underlying mechanisms of condensin. However, this loop extrusion on a bare DNA answers only a part of the whole puzzle of 3D organization of chromosome
We are interested in answering many other parts of the puzzle like: i) Are there more complex higher order loops?, ii) How do loops interact with each other?, iii) How do DNA loops interact with DNA supercoils (plectonemes) iii) What happens when DNA loops encounter nucleosome, RNA polymerase which are essential to the DNA structure and gene expression. We try to answer these questions by single-molecule techniques like magnetic tweezers and TIRFM which helps us explore the action of condensin motor with unprecedented details.
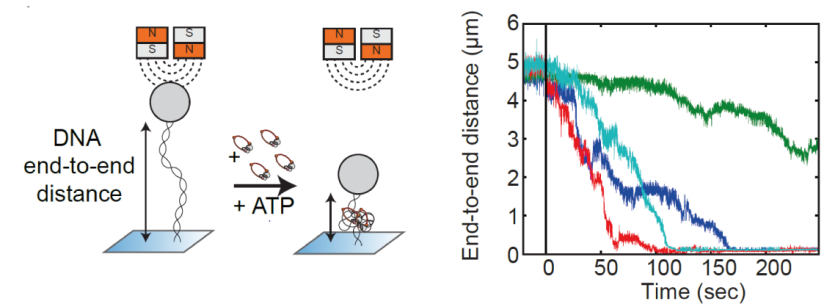
Fig.1. Real-time observation of condensin-driven DNA compaction upon ATP hydrolysis.

Movie.2 Visualization of a DNA loop that is extruded by condensin.
Direct visualization of conformational dynamics of SMC proteins
We are studying the structure and molecular mechanism of SMC proteins using atomic force microscopy (AFM). We succeeded to visualize condensin-mediated DNA loop to understand how it extrudes the DNA loop. Also, we visualized different conformations of condensin at the stem of loop. In addition, using a new, state of the art single-molecule technique called high speed atomic force microscopy (HS-AFM), we succeeded to obtain a movie of dynamical conformational changes of condensin. HS-AFM is an excellent technique for understanding protein’s structure and function relationship because it allows to directly visualize a protein’s structural dynamics with a high spatio-temporal resolution in a liquid phase. We showed that SMC arms are highly flexible and dynamic. Nowadays, we are expanding our approaches toward other SMC proteins.
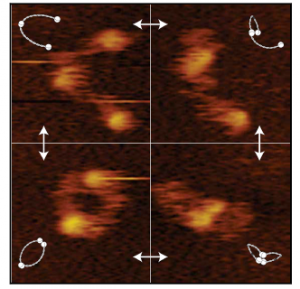
Figure 2. HS AFM video images on SMC dimers of condensin. The dynamical conformational changes show that the coiled-coils of SMC dimers are flexible and show extensive fluctuations in time (Eeftens et al. 2016, Cell Reports).
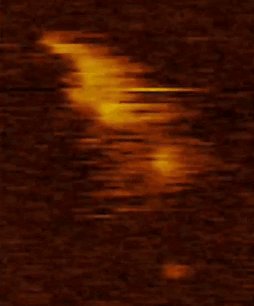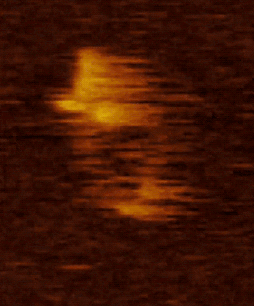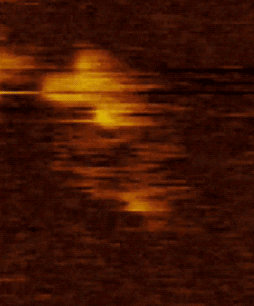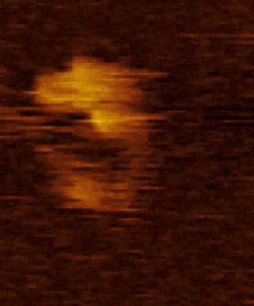
Movie 2. HS AFM video images of condensin with all subunits.

People working on this project

Eugene Kim

Je-Kyung Ryu
Mahipal Ganji

Biswajit Pradhan

Jacob W.J. Kerssemakers
- Room F0.110
- +31-(0)15-27 88331
- J.W.J.Kerssemakers@[REMOVE THIS]tudelft.nl
- Expertise: Single-molecule instrumentation

Jaco van der Torre
- Room F0.190
- +31-(0)15-27 83959
- J.vanderTorre@[REMOVE THIS]tudelft.nl
- Expertise: Molecular biology

Eli van der Sluis
- Room F0.130
- +31-(0)15-27 86572
- E.O.vandersluis@[REMOVE THIS]tudelft.nl
- Expertise: Protein purification

Allard Katan
- Room F0.190
- +31-(0)15-27 84561
- A.J.Katan@[REMOVE THIS]tudelft.nl
- Expertise: Scanning probe microscopy

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV









