Double nanopores
We are developing a novel mechanistic approach for manipulating single DNA molecules in the nanopore sensor (see our paper Pud et al, Nano Lett. 16, 8021 (2016)). The key idea is balancing the forces acting on DNA by stretching it in a nanoscale tug-of-war between two solid-state nanopores (Figure 1). This approach is promising to enrich solid-state nanopore platform with a single-molecule manipulation modality, which will enable it for DNA sequencing and studying DNA-protein interactions.
Our research in this direction is focused on developing chip-based double-nanopore devices (Figure 2) and using dual-barrel microcapillaries (in collaboration with Joshua Edel and Alexandar Ivanov labs) to control and manipulate DNA molecule during translocation (see our joint paper Cadinu et al, Nano Lett. 18 2738 (2018)).
Dual-barrel microcapillaries are easy to fabricate and they inherently enable the three-terminal geometry of the experiment (Figure 3). Each of the two nanopores has a separate electrode connected to an individual headstage of a dual-channel amplifier. The double-barrel experiment can be run in two modes of operation: competition and transfer mode. In the competition mode a positive bias is applied to both barrels. DNA molecules are attracted to each of the pores and thus there is a non-zero chance of single molecule getting trapped between them. We show that at higher voltages the probability of such events compared to regular single pore translocations can be up to 50%. The transfer mode is enabled when the pores are biased with opposite voltages, thus enabling DNA molecules to be expelled from one pore and captured into the second one. Depending on the pore biases the transfer efficiency can reach up to 100%.
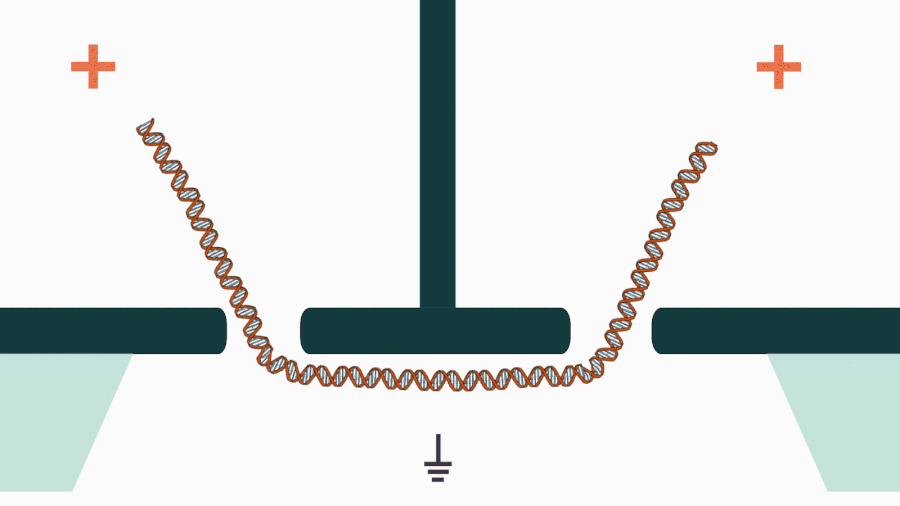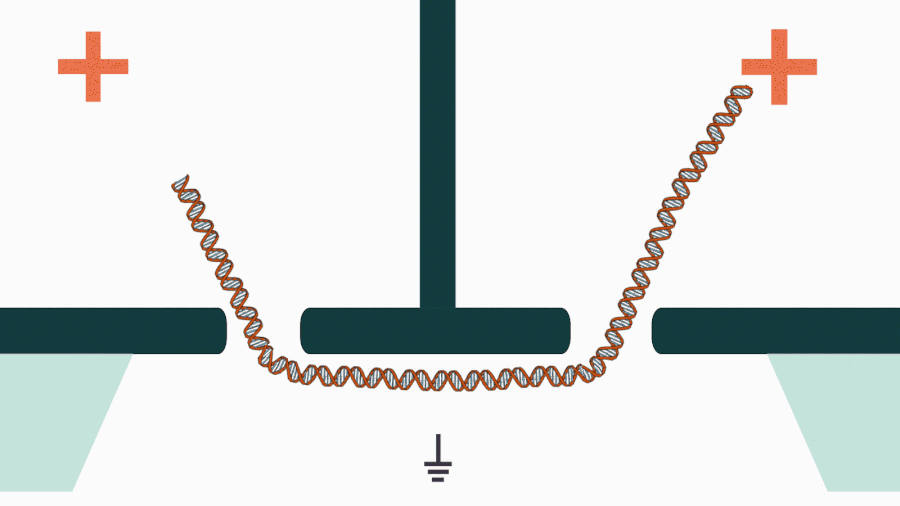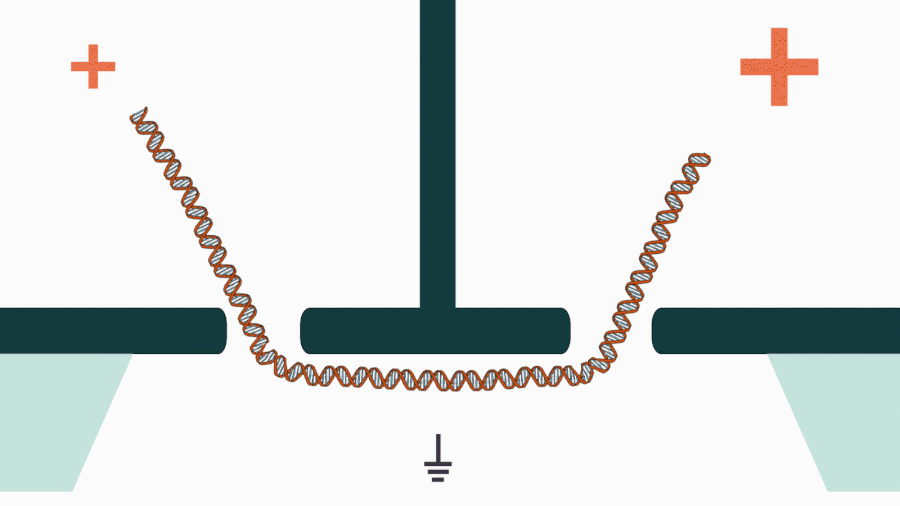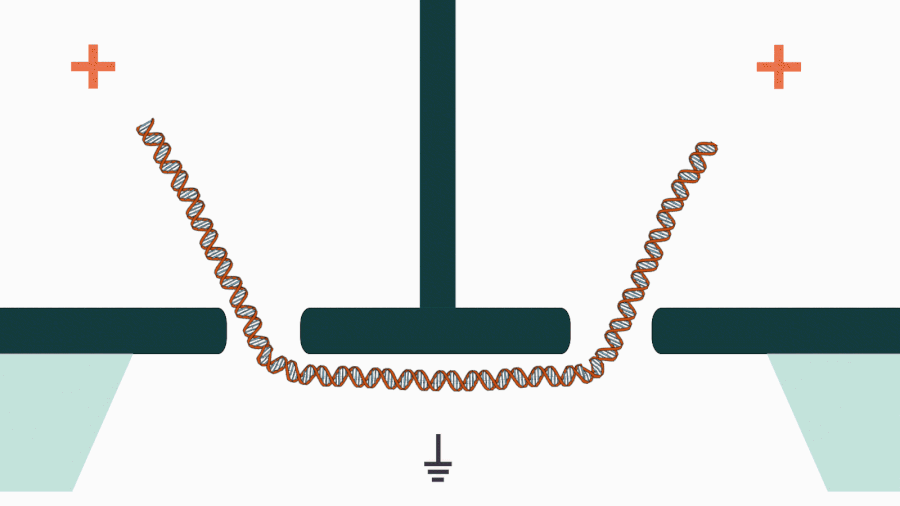
Figure 1. Concept of a controlled DNA movement in a double-nanopore system.
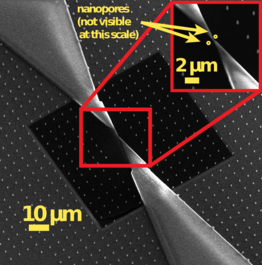
Figure 2. Dual nanopore chip with a nanoscale separator manufactured at our Kavli Nanolab @ Delft.
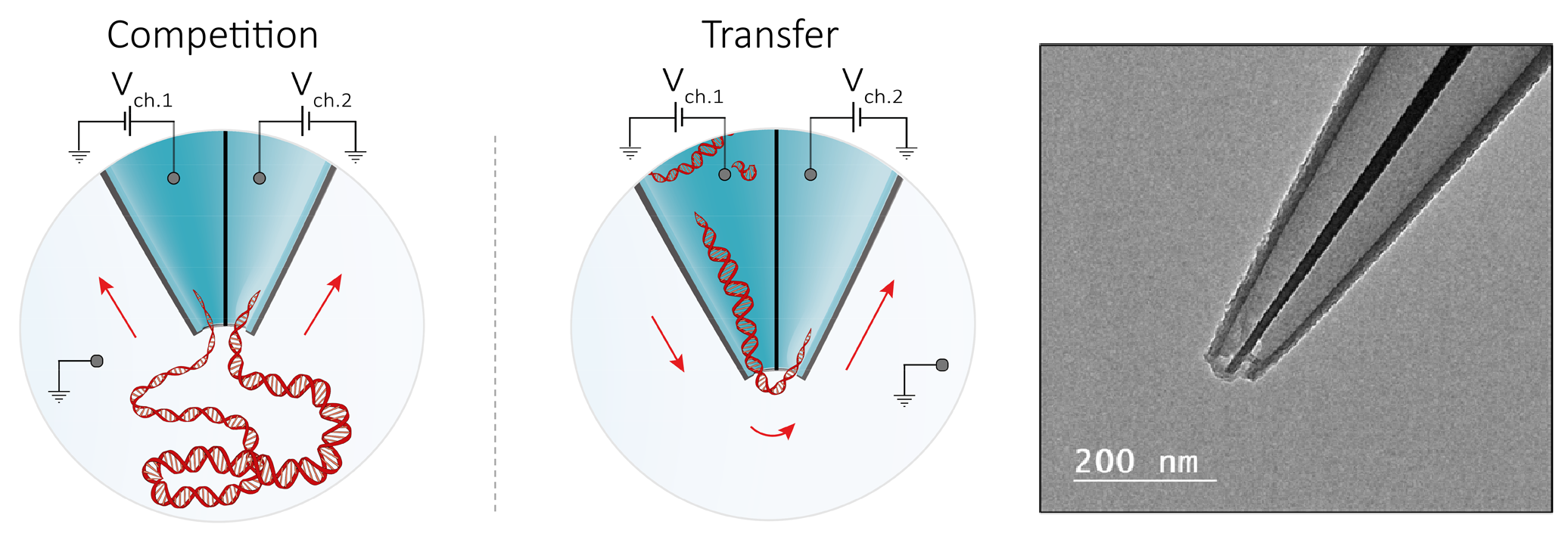
Figure 3. Left: Schematic representation of the double barrel nanopore experiment. Right: example of a double barrel nanopore.
People working on this project

Wayne Yang

Pinyao He
Sergii Pud

Cees Dekker
- F0.210
- +31-(0)15-27 86094
- C.Dekker@[REMOVE THIS]tudelft.nl
- Principal Investigator
- View CV


